4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
6O8S
 
 | | Syn-safencin 56 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
7RP3
 
 | | Crystal structure of GNE-1952 alkylated KRAS G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP2
 
 | | Crystal structure of Kas G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, GTPase KRas, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP4
 
 | | Crystal structure of KRAS G12C in complex with GNE-1952 | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
6VSP
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
6O8R
 
 | | Syn-safencin 24 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8J
 
 | | Syn-safencin | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8P
 
 | | Syn-safencin 8 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
6T2W
 
 | | Crystal structure of the CSF1R kinase domain with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZGZ
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
4YGX
 
 | |
4YH1
 
 | |
4YGY
 
 | |
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4GRB
 
 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
8C0P
 
 | |
8C0S
 
 | |
8CF3
 
 | |
6ILP
 
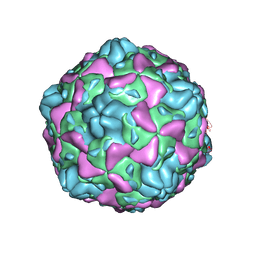 | | Cryo-EM structure of full Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6XYL
 
 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with L-serine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
6ILN
 
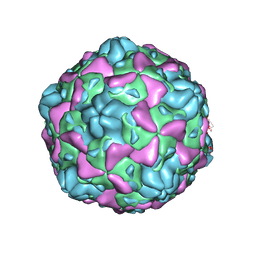 | | Cryo-EM structure of full Echovirus 6 particle at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
