3LKV
 
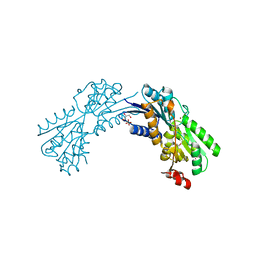 | | Crystal structure of conserved domain protein from vibrio cholerae o1 biovar eltor str. n16961 | | Descriptor: | GLYCEROL, PHENYLALANINE, SULFATE ION, ... | | Authors: | Nocek, B, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of conserved domain protein from vibrio cholerae o1 biovar eltor str. n16961
To be Published
|
|
1NI9
 
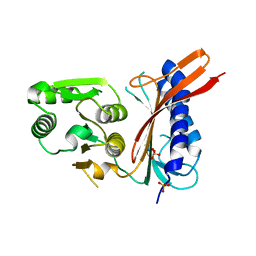 | | 2.0 A structure of glycerol metabolism protein from E. coli | | Descriptor: | Protein glpX, SULFATE ION | | Authors: | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-23 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3LUQ
 
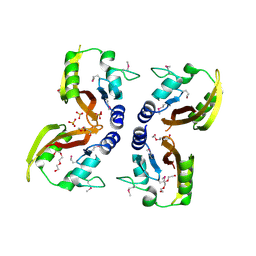 | | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A | | Descriptor: | SULFATE ION, Sensor protein, TRIETHYLENE GLYCOL | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A
To be Published
|
|
3IRA
 
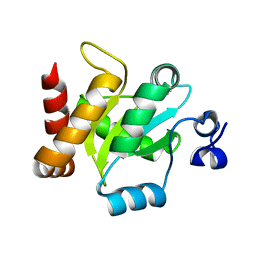 | |
3LVY
 
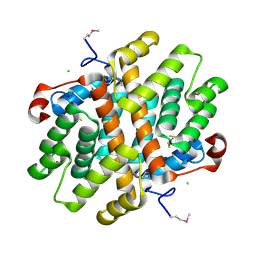 | | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Carboxymuconolactone decarboxylase family, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans
To be Published
|
|
3IH6
 
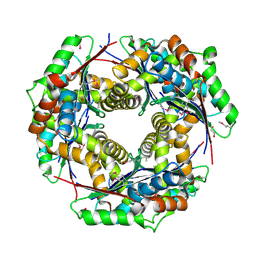 | | Crystal structure of putative zinc protease from Bordetella pertussis Tohama I | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative zinc protease | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative zinc protease from Bordetella pertussis Tohama I
To be Published
|
|
3I8N
 
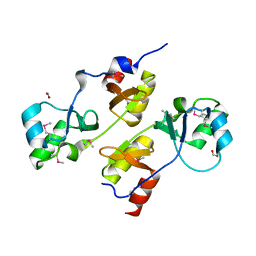 | |
3G7G
 
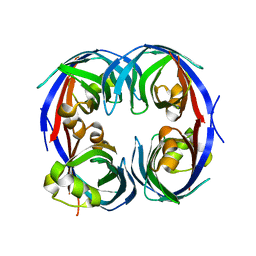 | |
3GE2
 
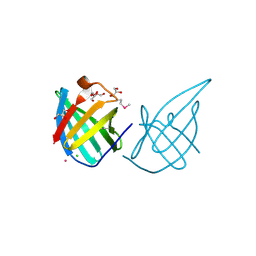 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
3G8W
 
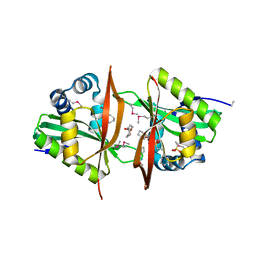 | | Crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, Lactococcal prophage ps3 protein 05 | | Authors: | Tan, K, Sather, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3QWT
 
 | |
1XEB
 
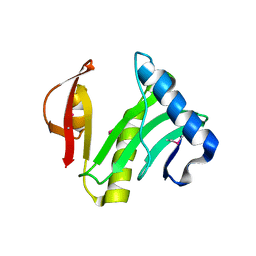 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA0115 | | Authors: | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
3MQO
 
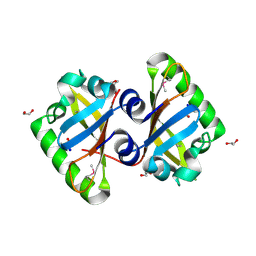 | | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A
To be Published
|
|
3UK0
 
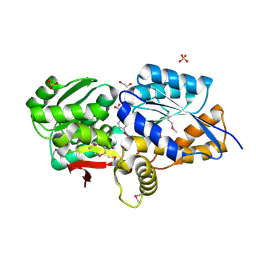 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3HL0
 
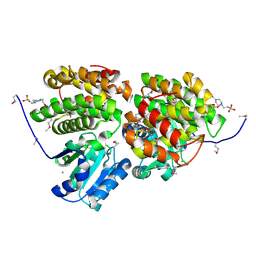 | | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Maleylacetate reductase, ... | | Authors: | Chang, C, Evdokimova, E, Mursleen, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens
To be Published
|
|
1L7A
 
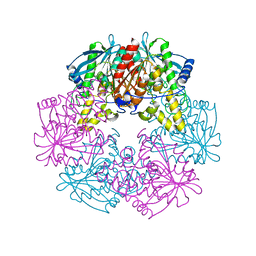 | |
3BJN
 
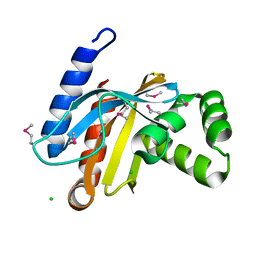 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3MKK
 
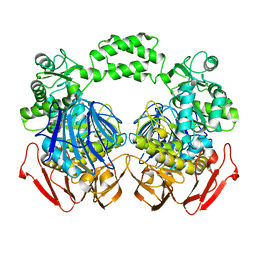 | | The crystal structure of the D307A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with isomaltose | | Descriptor: | alpha-D-glucopyranose, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, alpha-glucosidase GH31 family, ... | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structures Of The Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Faseb J., 24, 2010
|
|
3HLU
 
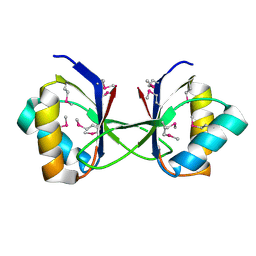 | |
3IV6
 
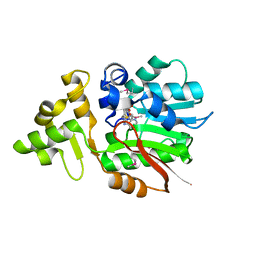 | | Crystal Structure of Putative Zn-dependent Alcohol Dehydrogenases from Rhodobacter sphaeroides. | | Descriptor: | CHLORIDE ION, Putative Zn-dependent Alcohol Dehydrogenase, S-ADENOSYLMETHIONINE | | Authors: | Kim, Y, Marshall, N, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Putative Zn-dependent Alcohol Dehydrogenases from Rhodobacter sphaeroides.
To be Published
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
3B4U
 
 | | Crystal structure of dihydrodipicolinate synthase from Agrobacterium tumefaciens str. C58 | | Descriptor: | Dihydrodipicolinate synthase, MAGNESIUM ION | | Authors: | Zhang, R, Xu, L, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-24 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the dihydrodipicolinate synthase from Agrobacterium tumefaciens.
To be Published
|
|
3MQQ
 
 | | The Crystal Structure of the PAS domain in complex with Ethanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.65A | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, SODIUM ION, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with Ethanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.65A
To be Published
|
|
3M6Y
 
 | | Structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at 1.45 a resolution. | | Descriptor: | 4-Hydroxy-2-oxoglutarate aldolase, CALCIUM ION, CHLORIDE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of 4-Hydroxy-2-Oxoglutarate Aldolase from Bacillus Cereus at 1.45 A Resolution.
To be Published
|
|
3SVI
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
