2ATZ
 
 | | Crystal structure of protein HP0184 from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, H. pylori predicted coding region HP0184 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of protein HP0184 from Helicobacter pylori
To be Published
|
|
1LSL
 
 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | Descriptor: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | Deposit date: | 2002-05-17 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
2EVV
 
 | | Crystal Structure of the PEBP-like Protein of Unknown Function HP0218 from Helicobacter pylori | | Descriptor: | GLYCEROL, SULFATE ION, hypothetical protein HP0218 | | Authors: | Kim, Y, Xu, X, Hong, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Hypothetical Protein HP0218 from Helicobacter pylori
To be Published
|
|
2F6S
 
 | | Structure of cell filamentation protein (fic) from Helicobacter pylori | | Descriptor: | PHOSPHATE ION, ZINC ION, cell filamentation protein, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-29 | | Release date: | 2006-01-10 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of cell filamentation protein (fic) from Helicobacter pylori
To be Published
|
|
1M3S
 
 | | Crystal structure of YckF from Bacillus subtilis | | Descriptor: | Hypothetical protein yckf | | Authors: | Sanishvili, R, Wu, R, Kim, D.E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-28 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Bacillus subtilis YckF: structural and functional evolution.
J.Struct.Biol., 148, 2004
|
|
2FA8
 
 | | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens | | Descriptor: | hypothetical protein Atu0228 | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-07 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens
To be Published, 2006
|
|
2FIU
 
 | | Crystal Structure of the Conserved Protein of Unknown Function ATU0297 from Agrobacterium tumefaciens | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Atu0297 from Agrobacterium tumefaciens
To be Published
|
|
2FIY
 
 | | The crystal structure of the FdhE protein from Pseudomonas aeruginosa | | Descriptor: | FE (III) ION, Protein fdhE homolog | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the FdhE protein from Pseudomonas aeruginosa PAO1
To be Published
|
|
2FD5
 
 | | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | transcriptional regulator | | Authors: | Zhang, R, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
4R1H
 
 | |
4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
4RBS
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | Descriptor: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | Authors: | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
4RAW
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Ampicillin
To be Published
|
|
4R7R
 
 | | Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | GLYCEROL, Putative lipoprotein | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To be Published
|
|
4R7U
 
 | | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Nocek, B, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin
To be Published
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | Descriptor: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
4R7Q
 
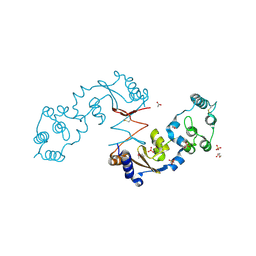 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4RCK
 
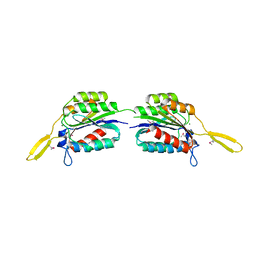 | |
4RDC
 
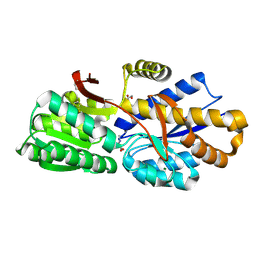 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline.
To be Published
|
|
4ICH
 
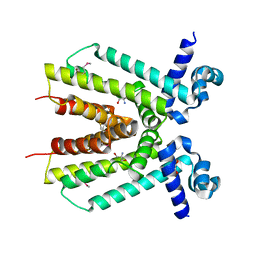 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
4R40
 
 | | Crystal Structure of TolB/Pal complex from Yersinia pestis. | | Descriptor: | FORMIC ACID, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of TolB/Pal complex from Yersinia pestis.
To be Published
|
|
4R6I
 
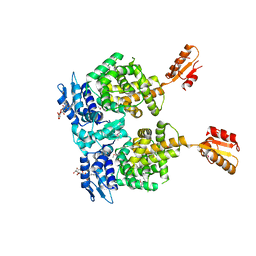 | | AtxA protein, a virulence regulator from Bacillus anthracis. | | Descriptor: | Anthrax toxin expression trans-acting positive regulator, DODECYL-BETA-D-MALTOSIDE | | Authors: | Osipiuk, J, Horton, L.B, Koehler, T.M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity.
Mol.Microbiol., 95, 2015
|
|
4RGI
 
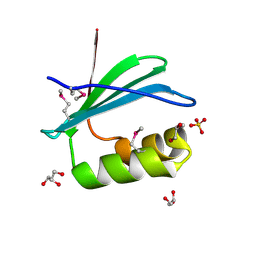 | | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis
To be Published
|
|
