7JYV
 
 | | Crystal Structure of HLA A*2402 in complex with YFSPIRVTF, an 9-mer influenza epitope | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYU
 
 | | Crystal Structure of HLA-A*2402 in complex with IYFSPIRVTF, an 10-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Rossjohn, J, Gras, S. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYW
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNW, an 9-mer influenza epitope | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, MHC class I antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYX
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNWET, an 11-mer epitope from Influenza | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PB2 peptide from Influenza, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7K0V
 
 | | Crystal structure of bRaf in complex with inhibitor GNE-0749 | | Descriptor: | CHLORIDE ION, N-(3,3-dimethylbutyl)-N'-{2-fluoro-5-[(5-fluoro-3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-methylphenyl}urea, Non-specific serine/threonine protein kinase | | Authors: | Yin, J, Eigenbrot, C.E, Wang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
7USE
 
 | | Cryo-EM structure of WAVE regulatory complex with Rac1 bound on both A and D site | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USC
 
 | | Cryo-EM structure of WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, Nck-associated protein 1, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USD
 
 | | Cryo-EM structure of D-site Rac1-bound WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
6SKA
 
 | | Teneurin 2 in complex with Latrophilin 1 Lec-Olf domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L1, ... | | Authors: | Chu, A, Carrasquero, M.A, Lowe, E, Seiradake, E. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons.
Cell, 180, 2020
|
|
6TBK
 
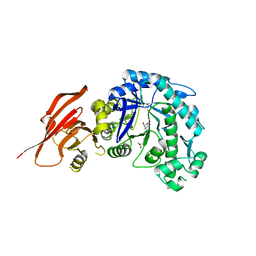 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{R},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6T3Z
 
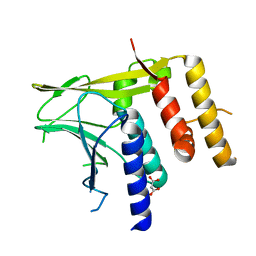 | | Crystal structure of the truncated EBV BFRF1-BFLF2 nuclear egress complex | | Descriptor: | MALONATE ION, Nuclear egress protein 2,Nuclear egress protein 1 | | Authors: | Muller, Y.A. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55862784 Å) | | Cite: | High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity.
J.Biol.Chem., 295, 2020
|
|
6THS
 
 | |
5KTU
 
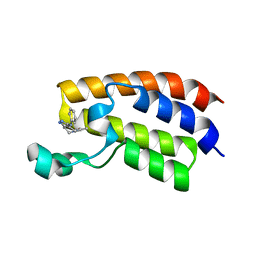 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | Descriptor: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
5WDO
 
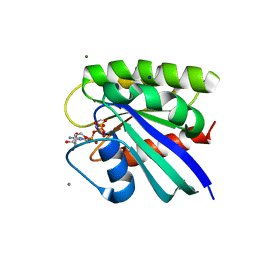 | | H-Ras bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5CTM
 
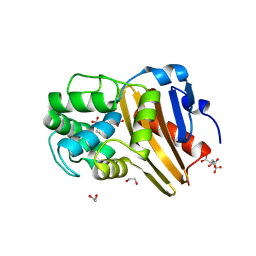 | | Structure of BPu1 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5EGS
 
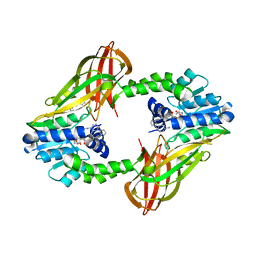 | | Human PRMT6 with bound fragment-type inhibitor | | Descriptor: | 2-[4-(phenylmethyl)piperidin-1-yl]ethanamine, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Steuber, H, Egner, U, Kania, J, Wu, H, Brown, P.J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Potent Class I Protein Arginine Methyltransferase Fragment Inhibitor.
J.Med.Chem., 59, 2016
|
|
7QBA
 
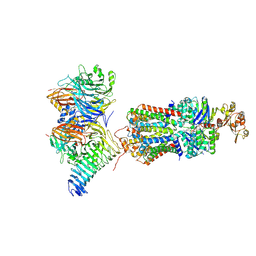 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
8AFP
 
 | | Structure of fibronectin 2 and 3 of L1CAM at 3.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neural cell adhesion molecule L1 | | Authors: | Guedez, G, Loew, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1.
Faseb J., 37, 2023
|
|
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
6TBJ
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{S},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TFI
 
 | | PXR IN COMPLEX WITH THROMBIN INHIBITOR COMPOUND 17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
5D5N
 
 | | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Complex | | Descriptor: | Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ZINC ION | | Authors: | Walzer, S.A, Egerer-Sieber, C, Hohl, K, Sevvana, M, Muller, Y.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions.
J.Biol.Chem., 290, 2015
|
|
6TDP
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GL dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDR
 
 | | Crystal structure of the disulfide engineered HLA-A0201 molecule devoid of peptide (annealed) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
5WDS
 
 | | Choanoflagellate Salpingoeca rosetta Ras with GDP bound | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
