7K6P
 
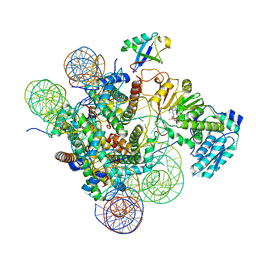 | | Active state Dot1 bound to the unacetylated H4 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7LV9
 
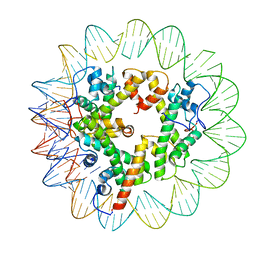 | | Marseillevirus heterotrimeric (hexameric) nucleosome | | Descriptor: | DNA (96-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LV8
 
 | | Structure of the Marseillevirus nucleosome | | Descriptor: | DNA (123-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1HFZ
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Pike, A.C.W, Brew, K, Acharya, K.R. | | Deposit date: | 1996-06-13 | | Release date: | 1997-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase.
Structure, 4, 1996
|
|
2FHH
 
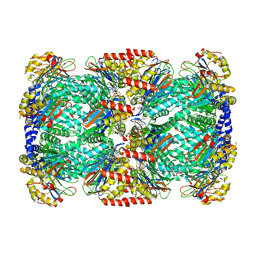 | |
8YE4
 
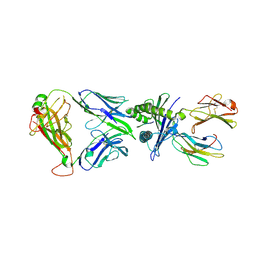 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
8ZV9
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
2Q57
 
 | |
4F23
 
 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
6KUU
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
7EVM
 
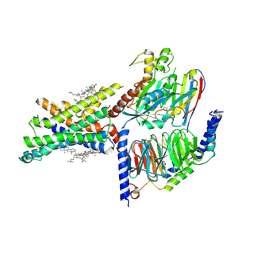 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
2H0D
 
 | |
5YAG
 
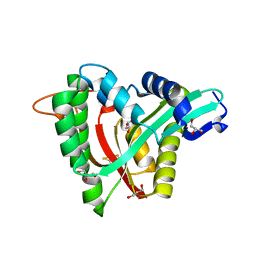 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | Descriptor: | AAEL004827-PA, GLYCEROL | | Authors: | Han, Q, Guan, H, Robinson, H, Li, J. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
6U3J
 
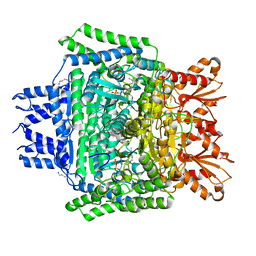 | | Structure of the 2-oxoadipate dehydrogenase DHTKD1 | | Descriptor: | 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, MAGNESIUM ION, ... | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex.
Acs Chem.Biol., 15, 2020
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | Descriptor: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0L
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
7C8P
 
 | |
