6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
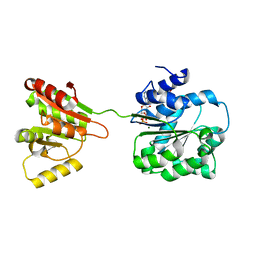 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
5IZ6
 
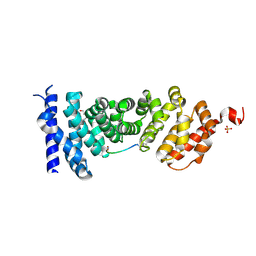 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5DJ5
 
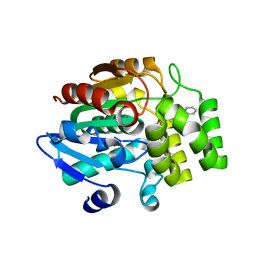 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
7FAH
 
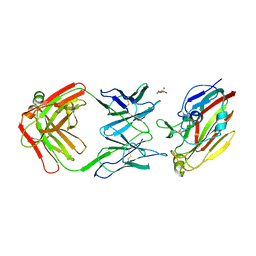 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
9J0K
 
 | | An agonist(compound 14e)of Thyroid Hormone Receptor B | | Descriptor: | 2-[[7-[3-(aminomethyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-08-02 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
9IX5
 
 | | An agonist(compound 15n) of Thyroid Hormone Receptor B | | Descriptor: | 2-[(1-methoxy-7-naphthalen-2-yloxy-4-oxidanyl-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-07-26 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
5DH5
 
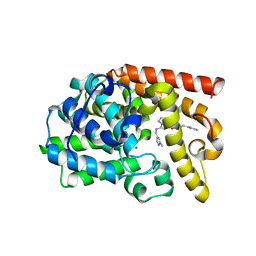 | | PDE10 complexed with N-[(1-methylpyrazol-4-yl)methyl]-5-[[(1S,2S)-2-(2-pyridyl)cyclopropyl]methoxy]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | MAGNESIUM ION, N-[(1-methyl-1H-pyrazol-4-yl)methyl]-5-{[(1S,2S)-2-(pyridin-2-yl)cyclopropyl]methoxy}pyrazolo[1,5-a]pyrimidin-7-amine, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8ZXV
 
 | | sweet protein MNEI-Mut 6-4 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
8ZXT
 
 | | sweet protein MNEI-Mut 6-3 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
8ZXY
 
 | | sweet protein MNEI-Mut 6-2 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
8ZXJ
 
 | | Sweet protein MNEI-Mut 6-1 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-14 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
7FCP
 
 | |
7FCQ
 
 | |
8HVS
 
 | |
7EW5
 
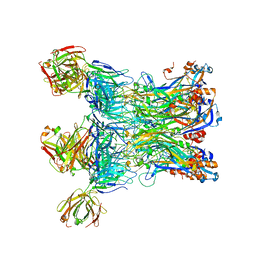 | | immune complex of HPV6 L1 pentamer and neutralizing antibody 13H5 | | Descriptor: | Heavy chain of 13H5, Light chain of 13H5, Major capsid protein L1 | | Authors: | Wang, Z.P, Wang, D.N, Gu, Y, Li, S.W. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Rational design of a cross-type HPV vaccine through immunodominance shift guided by a cross-neutralizing antibody.
Sci Bull (Beijing), 69, 2024
|
|
7F8I
 
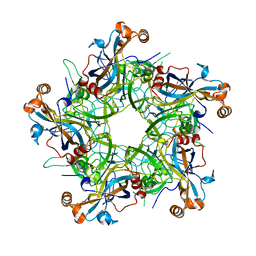 | | Crystal structure of HPV6 L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Wang, Z.P, Wang, D.N, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.366 Å) | | Cite: | Rational design of a cross-type HPV vaccine through immunodominance shift guided by a cross-neutralizing antibody.
Sci Bull (Beijing), 69, 2024
|
|
9ISF
 
 | |
9IVU
 
 | |
9IVT
 
 | |
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YPS
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5PGU
 
 | |
