4Y2M
 
 | | apo-GolB protein | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Zhao, J, Wang, F. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
6WK7
 
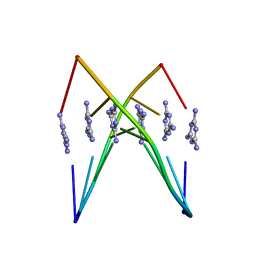 | | Crystal Structure Analysis of a poly(thymine) DNA duplex | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, DNA (5'-D(*TP*TP*TP*TP*TP*T)-3') | | Authors: | Li, Q, Zhao, J, Liu, L, Mandal, S, Rizzuto, F.J, He, H, Wei, S, Jonchhe, S, Sleiman, H.F, Mao, H, Mao, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | A poly(thymine)-melamine duplex for the assembly of DNA nanomaterials.
Nat Mater, 19, 2020
|
|
5GAR
 
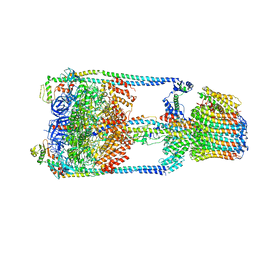 | | Thermus thermophilus V/A-ATPase, conformation 1 | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1MO0
 
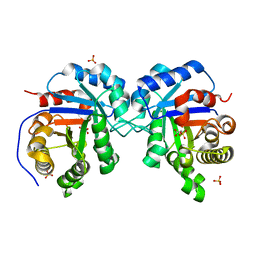 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
5HXD
 
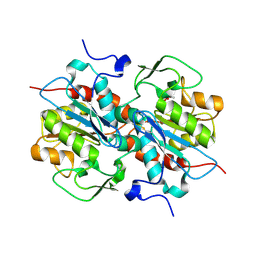 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | Descriptor: | CACODYLATE ION, Protein MpaA, ZINC ION | | Authors: | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
5I1M
 
 | | Yeast V-ATPase average of densities, a subunit segment | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7RUI
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844
To Be Published
|
|
7RUH
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844
To Be Published
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4DWT
 
 | |
4DWU
 
 | |
9IUZ
 
 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 187, 2024
|
|
6K61
 
 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
5DO2
 
 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
8GX2
 
 | | The crystal structure of human CtsL in complex with 14c | | Descriptor: | DIMETHYL SULFOXIDE, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
8TM4
 
 | | Human pre 13S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM5
 
 | | Human mixed 13S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM3
 
 | | Human proteasome alpha ring assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome assembly chaperone 3, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
8TM6
 
 | | Human premature 20S proteasome assembly intermediate | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Zhang, H, Zhao, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human 20S proteasome biogenesis.
Nat Commun, 15, 2024
|
|
5F0W
 
 | |
5F0U
 
 | | Crystal structure of Gold binding protein | | Descriptor: | Putative copper chaperone, SILVER ION | | Authors: | Wei, W, Wang, F, Zhao, J. | | Deposit date: | 2015-11-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of tetrasilver bound to GolB at 1.70 Angstroms resolution
To Be Published
|
|
