5D7D
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
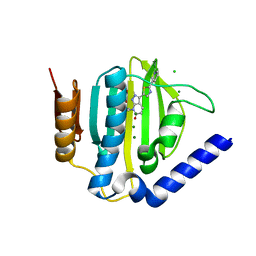
3X21
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
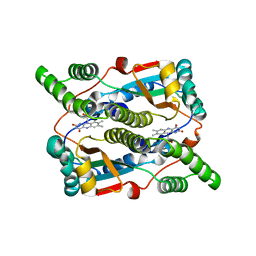
3WL1
 
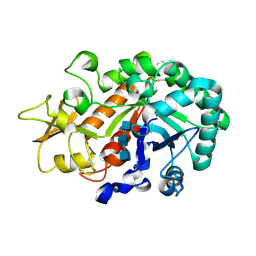 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WMB
 
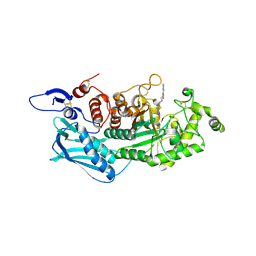 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | Descriptor: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3X22
 
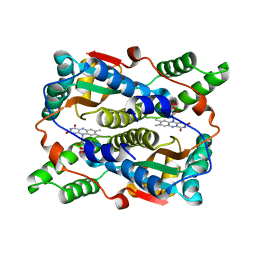 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
1J9I
 
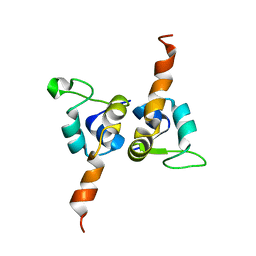 | | STRUCTURE OF THE DNA BINDING DOMAIN OF THE GPNU1 SUBUNIT OF LAMBDA TERMINASE | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | De Beer, T, Meyer, J, Ortega, M, Yang, Q, Maes, L, Duffy, C, Berton, N, Sippy, J, Overduin, M, Feiss, M, Catalano, C. | | Deposit date: | 2001-05-25 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into specific DNA recognition during the assembly of a viral genome packaging machine.
Mol.Cell, 9, 2002
|
|
3NSM
 
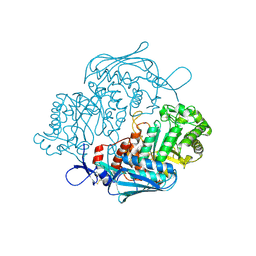 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3WQV
 
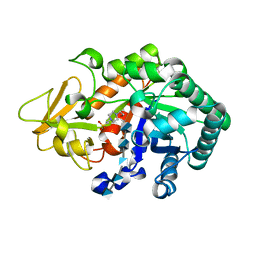 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with a(GlcN)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chen, L, Zhou, Y, Yang, Q. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WZF
 
 | | Crystal structure of human cytoplasmic aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Jiang, X, Chang, H, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2014-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Recombinant expression, purification and Preliminary crystallographic studies of human cytoplasmic aspartate aminotransferase
To be Published
|
|
3WL0
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKZ
 
 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W4R
 
 | | Crystal structure of an insect chitinase from the Asian corn borer, Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Shen, X, Yang, Q. | | Deposit date: | 2013-01-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YG7
 
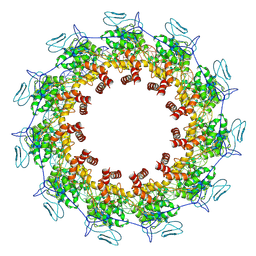 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
3WMC
 
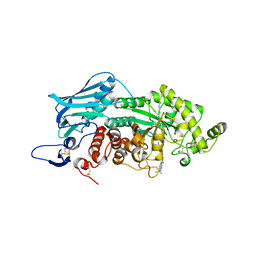 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | Authors: | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3OZO
 
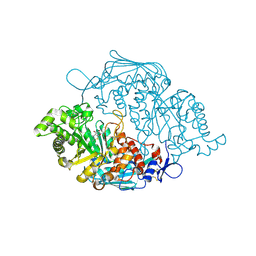 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
3OZP
 
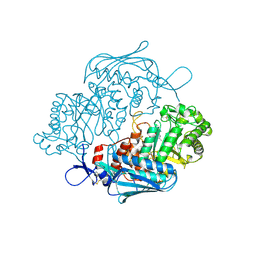 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases
Biochem.J., 438, 2011
|
|
5T0I
 
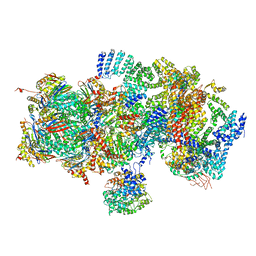 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0C
 
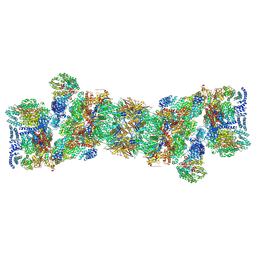 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0J
 
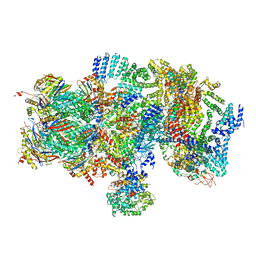 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0G
 
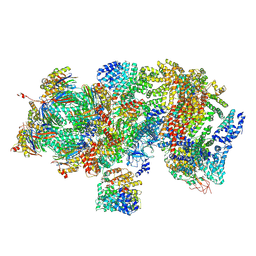 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0H
 
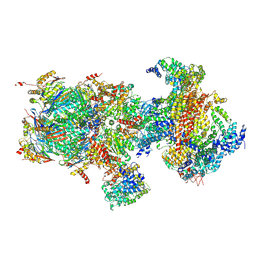 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3S6T
 
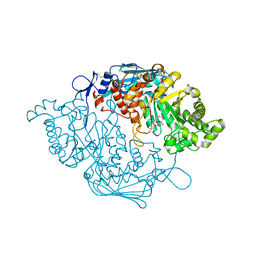 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Yang, Q, Shen, X. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
7X1Z
 
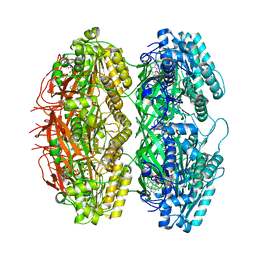 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
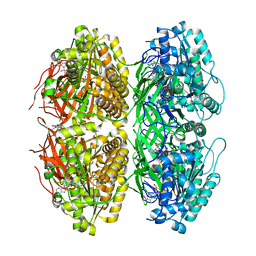 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
