4EEE
 
 | |
4EKX
 
 | |
7KSP
 
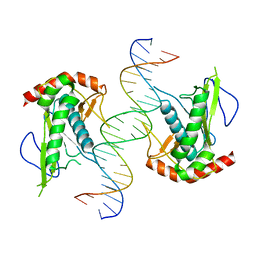 | | Crystal structure of hSAMD9_DBD with DNA | | Descriptor: | DNA, Sterile alpha motif domain-containing protein 9 | | Authors: | Peng, S, Pathak, P, Xiang, Y, Deng, J. | | Deposit date: | 2020-11-23 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of an effector domain in antiviral factors and tumor suppressors SAMD9 and SAMD9L.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4XFU
 
 | | Structure of IL-18 SER Mutant V | | Descriptor: | Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFT
 
 | | Structure of IL-18 SER Mutant III | | Descriptor: | DIMETHYL SULFOXIDE, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFS
 
 | | Structure of IL-18 SER Mutant I | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3MUU
 
 | | Crystal structure of the Sindbis virus E2-E1 heterodimer at low pH | | Descriptor: | Structural polyprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L, Jose, J, Xiang, Y, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2010-05-03 | | Release date: | 2010-11-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural changes of envelope proteins during alphavirus fusion.
Nature, 468, 2010
|
|
3MUW
 
 | | Pseudo-atomic structure of the E2-E1 protein shell in Sindbis virus | | Descriptor: | Structural polyprotein | | Authors: | Li, L, Jose, J, Xiang, Y, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2010-05-03 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural changes of envelope proteins during alphavirus fusion.
Nature, 468, 2010
|
|
5FB5
 
 | |
1X7I
 
 | | Crystal structure of the native copper homeostasis protein (cutCm) with calcium binding from Shigella flexneri 2a str. 301 | | Descriptor: | CALCIUM ION, Copper homeostasis protein cutC | | Authors: | Zhu, D.Y, Zhu, Y.Q, Huang, R.H, Xiang, Y, Wang, D.C. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the copper homeostasis protein (CutCm) from Shigella flexneri at 1.7 A resolution: The first structure of a new sequence family of TIM barrels
Proteins, 58, 2004
|
|
3J2Z
 
 | |
3J2Y
 
 | |
3JA7
 
 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
3J2W
 
 | | Electron cryo-microscopy of Chikungunya virus | | Descriptor: | Capsid protein, Glycoprotein E1, Glycoprotein E2 | | Authors: | Sun, S, Xiang, Y, Rossmann, M.G. | | Deposit date: | 2013-01-28 | | Release date: | 2013-04-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
3J2X
 
 | |
3J30
 
 | |
5FEI
 
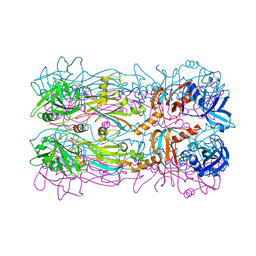 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
5FB4
 
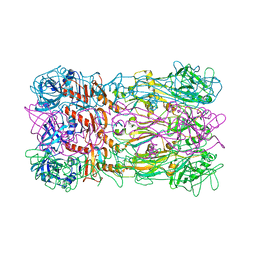 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
6A7F
 
 | |
7K39
 
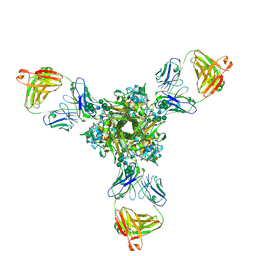 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3A
 
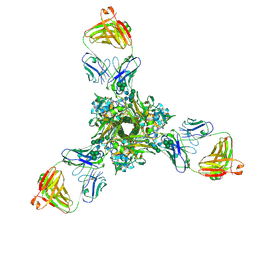 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3B
 
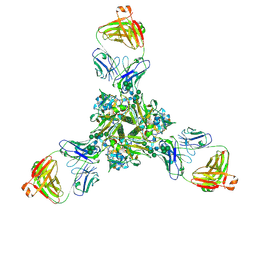 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K37
 
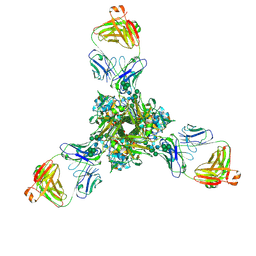 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
3TXS
 
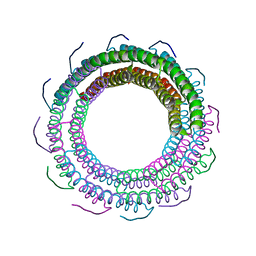 | | Crystal Structure of phage 44RR small terminase gp16 | | Descriptor: | Terminase DNA packaging enzyme small subunit | | Authors: | Sun, S, Xiang, Y, Rossmann, M.G. | | Deposit date: | 2011-09-23 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
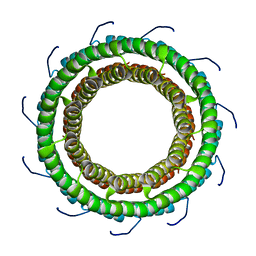
3TXQ
 
 | |
