7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
8JFV
 
 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
8S5B
 
 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
6S5J
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-Ethyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-ethyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5Q
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-isobutyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-(2-methylpropyl)-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5U
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with N-[2-(1H-Indol-3-yl)ethyl]-3-methyl-1-butanamine | | Descriptor: | Strictosidine synthase, ~{N}-[2-(1~{H}-indol-3-yl)ethyl]-3-methyl-butan-1-amine | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5M
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-n-propyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-propyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
4UN2
 
 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5A3G
 
 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KSP
 
 | | Mechanism for the selective interaction of C-terminal EH-domain proteins with specific NPF-containing partners | | Descriptor: | CALCIUM ION, EH domain-containing protein 1, MICAL L1 like peptide | | Authors: | Kieken, F, Sharma, M, Jovic, M, Giridharan, S.S, Naslavsky, N, Caplan, S, Sorgen, P.L. | | Deposit date: | 2010-01-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners.
J.Biol.Chem., 285, 2010
|
|
7BY6
 
 | | Plasmodium vivax cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD1389 | | Descriptor: | (3S,4R,8R,9R,10S)-N-(4-cyclopropyloxyphenyl)-10-(methoxymethyl)-3,4-bis(oxidanyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Malhotra, N, Manmohan, S, Harlos, K, Melillo, B, Schreiber, S.L, Manickam, Y, Sharma, S. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural basis of malaria parasite phenylalanine tRNA-synthetase inhibition by bicyclic azetidines.
Nat Commun, 12, 2021
|
|
7DPI
 
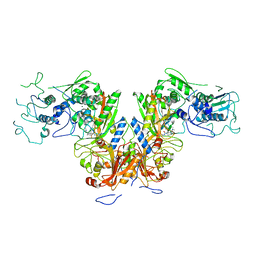 | | Plasmodium falciparum cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD7929 | | Descriptor: | (8R,9S,10S)-10-[(dimethylamino)methyl]-N-(4-methoxyphenyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanine--tRNA ligase, ... | | Authors: | Manmohan, S, Malhotra, N, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2020-12-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.597 Å) | | Cite: | Inhibition of Plasmodium falciparum phenylalanine tRNA synthetase provides opportunity for antimalarial drug development.
Structure, 30, 2022
|
|
6NF8
 
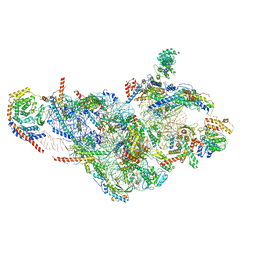 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | Descriptor: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NEQ
 
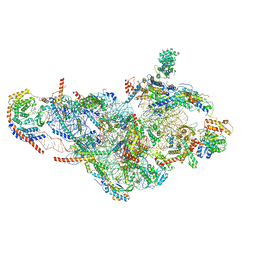 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | Descriptor: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
5Z79
 
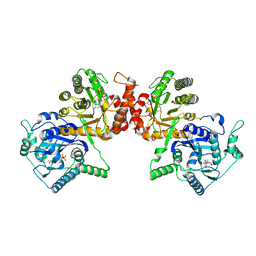 | | Crystal Structure Analysis of the HPPK-DHPS in complex with substrates | | Descriptor: | 4-AMINOBENZOIC ACID, 5'-DEOXYADENOSINE, 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, ... | | Authors: | Manickam, Y, Karl, H, Sharma, A. | | Deposit date: | 2018-01-27 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase-dihydropteroate synthase fromPlasmodium vivaxsheds light on drug resistance
J. Biol. Chem., 293, 2018
|
|
6IPA
 
 | |
7FHR
 
 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mahto, J.K, Dhankhar, P, Kumar, P. | | Deposit date: | 2021-07-30 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
7FJL
 
 | |
7V25
 
 | |
7V28
 
 | |
7VJU
 
 | |
7QHV
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with sulfoquinovosyl diacylglycerol | | Descriptor: | GLYCINE, Sulfoquinovosyl binding protein, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-[(2~{S})-3-butanoyloxy-2-heptanoyloxy-propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A, Davies, G.J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
8JFF
 
 | |
7YZS
 
 | |
