4DZH
 
 | | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn
to be published
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
4F0S
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine. | | Descriptor: | 5-methylthioadenosine/S-adenosylhomocysteine deaminase, CHLORIDE ION, INOSINE, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine.
To be Published
|
|
3N2C
 
 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3O7U
 
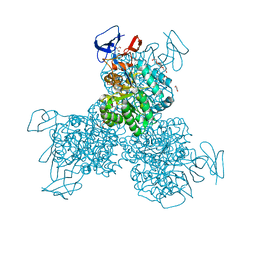 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
3PNZ
 
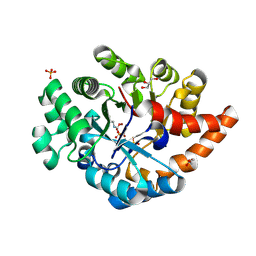 | | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphotriesterase family protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-11-20 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.5983 Å) | | Cite: | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes
To be Published
|
|
3RHG
 
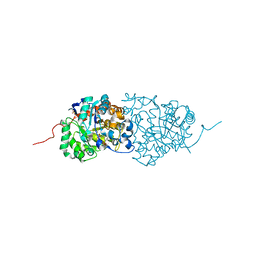 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | Descriptor: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3R0D
 
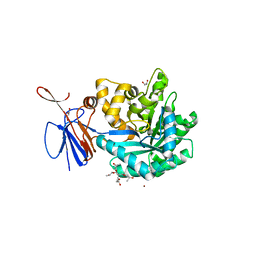 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Kamat, S, Hitchcock, D, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-03-07 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site
To be Published
|
|
3RN6
 
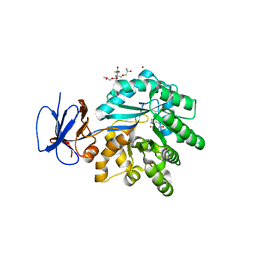 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and isoguanine | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 6-amino-3,7-dihydro-2H-purin-2-one, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Rescue of the orphan enzyme isoguanine deaminase.
Biochemistry, 50, 2011
|
|
3S2L
 
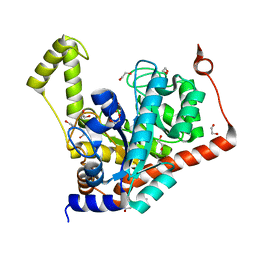 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Glu | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Glu
To be Published
|
|
3RYS
 
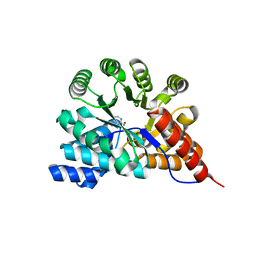 | |
3S2J
 
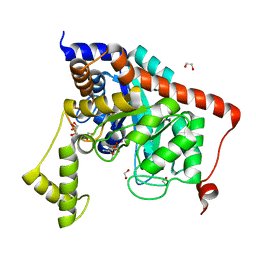 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Ala
To be Published
|
|
3S2N
 
 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Tyr-D-Asp | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-2-(4-hydroxyphenyl)ethyl](hydroxy)phosphoryl]methyl}butanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Tyr-D-Asp
To be Published
|
|
3S2M
 
 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Phe-D-Asp | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]methyl}butanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Phe-D-Asp
To be Published
|
|
4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4NI8
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-carboxyvanillate decarboxylase, 5-methoxybenzene-1,3-dicarboxylic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid
To be Published
|
|
4NG3
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid
To be Published
|
|
4QRN
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE AND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-Carboxyvanillate Decarboxylase, ACETATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QS6
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID, NO METAL, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Ligw2 Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4R88
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine | | Descriptor: | 1,2-ETHANEDIOL, 5-fluorocytosine, ACETIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine
To be Published
|
|
4RDV
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
4QTG
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE | | Descriptor: | 5-CARBOXYVANILLATE DECARBOXYLASE, GLYCEROL, MANGANESE (II) ION | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase LIGW2 from Novosphingobium Aromaticivorans
To be Published
|
|
4QSF
 
 | | CRYSTAL STRUCTURE of AMIDOHYDROLASE PMI1525 (TARGET EFI-500319) FROM PROTEUS MIRABILIS HI4320, A COMPLEX WITH BUTYRIC ACID AND MANGANESE | | Descriptor: | Amidohydrolase Pmi1525, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Xiang, D.F, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
4R85
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine | | Descriptor: | 5-methylcytosine, Cytosine deaminase, FE (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine
To be Published
|
|
