1PY4
 
 | | Beta2 microglobulin mutant H31Y displays hints for amyloid formations | | Descriptor: | Beta-2-microglobulin precursor | | Authors: | Rosano, C, Zuccotti, S, Mangione, P, Giorgetti, S, Bellotti, V, Pettirossi, F, Corazza, A, Viglino, P, Esposito, G, Bolognesi, M. | | Deposit date: | 2003-07-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | beta2-microglobulin H31Y variant 3D structure highlights the protein natural propensity towards intermolecular aggregation
J.Mol.Biol., 335, 2004
|
|
4JC4
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2013-02-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
1UWW
 
 | | X-ray crystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. | | Descriptor: | CALCIUM ION, ENDOGLUCANASE | | Authors: | Jamal, S, Nurizzo, D, Boraston, A, Davies, G.J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Crystal Structure of a Non-Crystalline Cellulose-Specific Carbohydrate-Binding Module: Cbm28
J.Mol.Biol., 339, 2004
|
|
2OII
 
 | | Structure of EMILIN-1 C1q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Colebrooke, S.A, Corazza, A, Cicero, D.O, Eliseo, T, Viglino, P, Campbell, I.D, Colombatti, A, Esposito, G. | | Deposit date: | 2007-01-11 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of EMILIN-1
To be Published
|
|
2GV1
 
 | |
3KK0
 
 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
3P2J
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Yadav, R, Sinha, M, Arora, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution
To be Published
|
|
2KVK
 
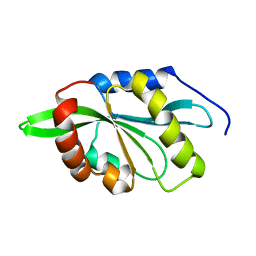 | | Solution structure of ADF/cofilin (LDCOF) from Leishmania donovani | | Descriptor: | Actin severing and dynamics regulatory protein | | Authors: | Pathak, P.P, Pulavarti, S.V, Jain, A, Sahasrabuddhe, A.A, Gupta, C.M, Arora, A. | | Deposit date: | 2010-03-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF/cofilin from Leishmania donovani
J.Struct.Biol., 172, 2010
|
|
3EZR
 
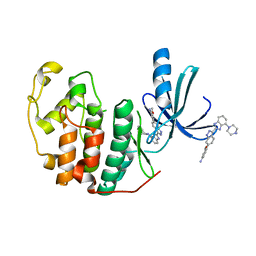 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3F5X
 
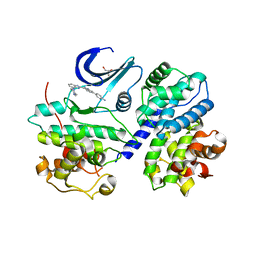 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EZV
 
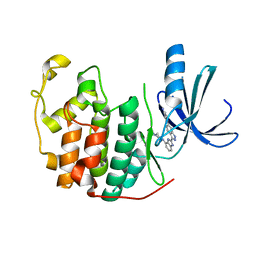 | | CDK-2 with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5O5X
 
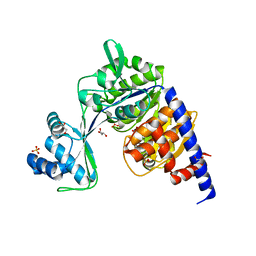 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Y
 
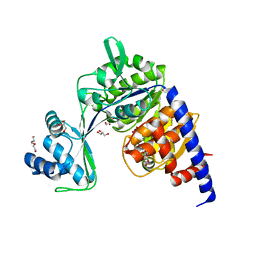 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
2NAF
 
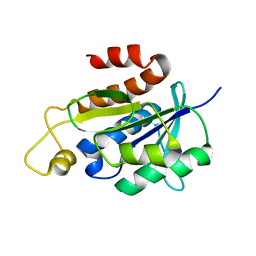 | | Solution structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Fatma, F, Kabra, A, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2015-12-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of peptidyl-tRNA hydrolase from Mycobacterium smegmatis by NMR spectroscopy.
Biochim.Biophys.Acta, 1864, 2016
|
|
2LBU
 
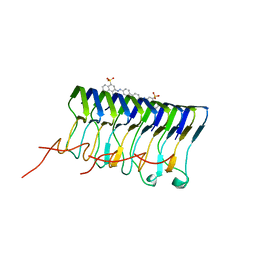 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | Descriptor: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
8IU6
 
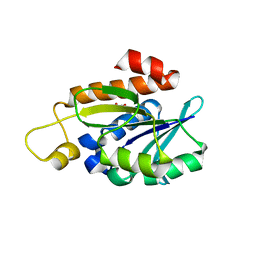 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
2LRA
 
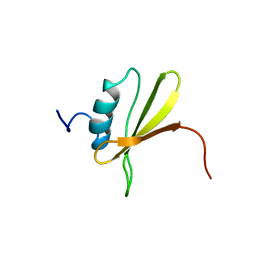 | | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S, Yadav, R, Jain, A, Pathak, P, Meher, A, Arora, A. | | Deposit date: | 2012-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag
To be Published
|
|
2LXX
 
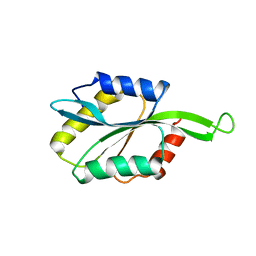 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 2, isoform c | | Authors: | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
8JQU
 
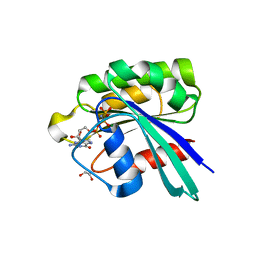 | | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Pandey, D, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani
To Be Published
|
|
8JXK
 
 | | Crystal Structure of Rv0047c from Mycobacterium tuberculosis | | Descriptor: | Conserved protein | | Authors: | Ansari, M.S, Yadav, V, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Rv0047c from Mycobacterium tuberculosis
To Be Published
|
|
2MP4
 
 | | Solution Structure of ADF like UNC-60A Protein of Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 1, isoforms a/b | | Authors: | Shukla, V, Yadav, R, Kabra, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone dynamics of ADF like UNC-60A protein from Caenorhabditis elegans: its divergence from conventional ADF/cofilin
To be Published
|
|
2JRC
 
 | |
2LGJ
 
 | | Solution structure of MsPTH | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
4EDZ
 
 | | Crystal structure of hH-PGDS with water displacing inhibitor | | Descriptor: | 4-(3-methylisoquinolin-1-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Day, J.E, Thorarensen, A, Trujillo, J.I. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the binding pocket of human hematopoietic prostaglandin (PG) D2 synthase (hH-PGDS): a tale of two waters.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
