8B18
 
 | | DtpB-Nb132-AF | | Descriptor: | ALA-PHE, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1C
 
 | | DtpB-Nb132-ALA | | Descriptor: | ALA-LEU-ALA, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B19
 
 | | DtpB-Nb132-AFA | | Descriptor: | ALA-PHE-ALA, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B17
 
 | | DtpB-Nb132-AWA | | Descriptor: | ALA-TRP-ALA, DECANE, DODECANE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
8B1H
 
 | | DtpB-Nb132-KV | | Descriptor: | DECANE, DODECANE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Killer, M, Finocchio, G, Lei, J, Jungnickel, K, Kotov, V, Steinke, J, Bartels, K, Strauss, J, Dupeux, F, Humm, A.S, Cornaciu, I, Marquez, J, Pardon, E, Steyeart, J, Loew, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Cell Rep, 42, 2023
|
|
7BJS
 
 | | Crystal structure of Khc/atypical Tm1 complex | | Descriptor: | Kinesin heavy chain, SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJN
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 270-334 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJG
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 262-363 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
6GHJ
 
 | | PepTSt in complex with tripeptide Phe-Ala-Gln | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-05-08 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Tripeptide binding in a proton-dependent oligopeptide transporter.
FEBS Lett., 592, 2018
|
|
6GS7
 
 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
4DT4
 
 | |
4K25
 
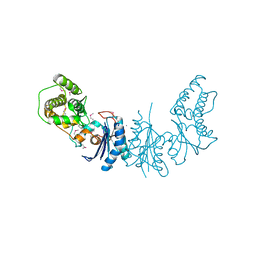 | | Crystal Structure of yeast Qri7 homodimer | | Descriptor: | CALCIUM ION, Probable tRNA threonylcarbamoyladenosine biosynthesis protein QRI7, mitochondrial, ... | | Authors: | Neculai, D, Wan, L, Mao, D.Y, Sicheri, F. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system.
Nucleic Acids Res., 41, 2013
|
|
8BJ6
 
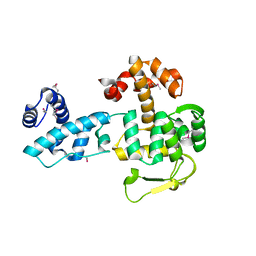 | | Crystal structure of YopR | | Descriptor: | SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a unique repression system present in arbitrium phages of the SPbeta family.
Cell Host Microbe, 31, 2023
|
|
5OXP
 
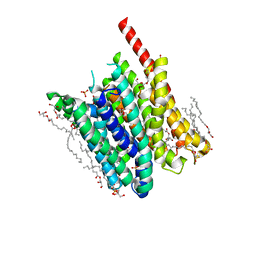 | | PepTSt in occluded conformation with phosphate ion bound | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXL
 
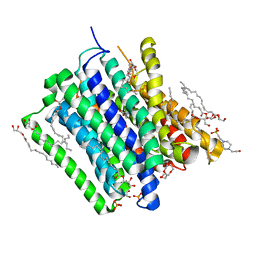 | | PepTSt in complex with dipeptide Ala-Leu | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXM
 
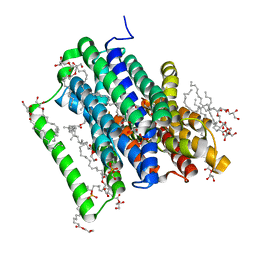 | | PepTSt in complex with dipeptide Asp-Glu | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ASPARTIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXK
 
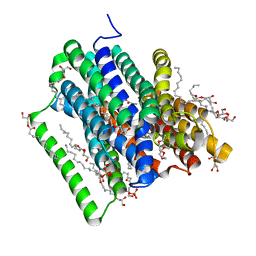 | | PepTSt in complex with dipeptide Ala-Gln | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ALANINE, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXO
 
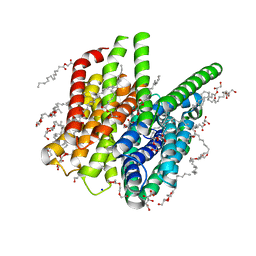 | | PepTSt apo structure | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, CITRIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXQ
 
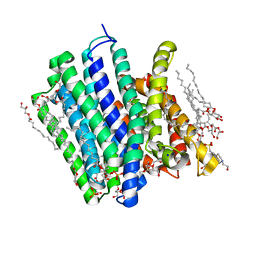 | | PepTSt in complex with HEPES (300 mM) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5OXN
 
 | | PepTSt in complex with dipeptide Phe-Ala | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ALANINE, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
3FT7
 
 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
2F52
 
 | |
1Z1L
 
 | | The Crystal Structure of the Phosphodiesterase 2A Catalytic Domain | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Ding, Y.H, Kohls, D, Low, C. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants for Inhibitor Specificity and Selectivity in PDE2A Using the Wheat Germ in Vitro Translation System.
Biochemistry, 44, 2005
|
|
2K9I
 
 | |
6FNE
 
 | | Structure of human Brag2 (Sec7-PH domains) with the inhibitor Bragsin bound to the PH domain | | Descriptor: | (2~{S})-6-methyl-5-nitro-2-(trifluoromethyl)-2,3-dihydrochromen-4-one, IQ motif and SEC7 domain-containing protein 1, NONAETHYLENE GLYCOL | | Authors: | Nawrotek, A, Zeghouf, M, Cherfils, J. | | Deposit date: | 2018-02-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling.
Nat.Chem.Biol., 15, 2019
|
|
