3NTW
 
 | |
5TDC
 
 | | Crystal structure of the human UBR-box domain from UBR1 in complex with monomethylated arginine peptide. | | Descriptor: | E3 ubiquitin-protein ligase UBR1, NMM-ILE-PHE-SER peptide, SULFATE ION, ... | | Authors: | Kozlov, G, Munoz-Escobar, J, Matta-Camacho, E, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TSR
 
 | |
3RG0
 
 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
1GH9
 
 | |
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5K23
 
 | |
4NWY
 
 | |
7M1U
 
 | | Crystal structure of an archaeal CNNM, MtCorB, R235L mutant with C-terminal deletion | | Descriptor: | Hemolysin, contains CBS domains | | Authors: | Chen, Y.S, Kozlov, G, Gehring, K. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
7M1T
 
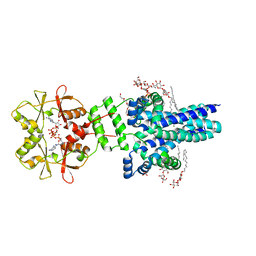 | | Crystal structure of an archaeal CNNM, MtCorB, with C-terminal deletion in complex with Mg2+-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hemolysin, contains CBS domains, ... | | Authors: | Chen, Y.S, Kozlov, G, Gehring, K. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
8CT8
 
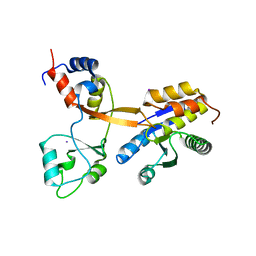 | | Crystal structure of Drosophila melanogaster PRL/CBS-pair domain complex | | Descriptor: | IODIDE ION, PRL-1 phosphatase, Unextended protein | | Authors: | Fakih, R, Goldstein, R.H, Kozlov, G, Gehring, K. | | Deposit date: | 2022-05-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Burst kinetics and CNNM binding are evolutionarily conserved properties of phosphatases of regenerating liver.
J.Biol.Chem., 299, 2023
|
|
4K95
 
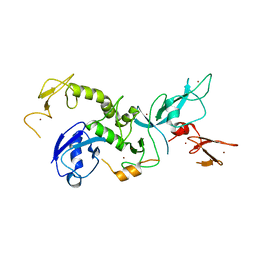 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4JU5
 
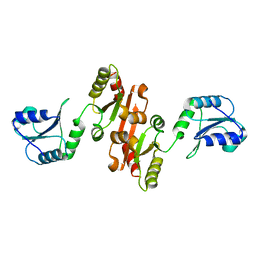 | |
4JRD
 
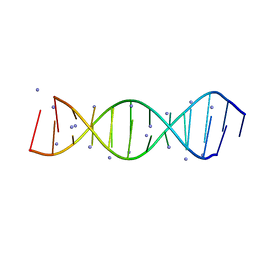 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1NMR
 
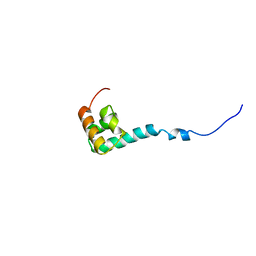 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
5K35
 
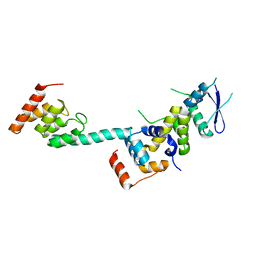 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
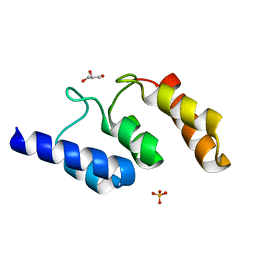 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K25
 
 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5TDA
 
 | |
3IQL
 
 | | Crystal structure of the rat endophilin-A1 SH3 domain | | Descriptor: | CHLORIDE ION, Endophilin-A1 | | Authors: | Trempe, J.F, Kozlov, G, Camacho, E.M, Gehring, K. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
5VMD
 
 | | Crystal structure of UBR-box from UBR6 in a domain-swapping conformation | | Descriptor: | 1,2-ETHANEDIOL, F-box only protein 11, ZINC ION | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Protein Sci., 26, 2017
|
|
5V44
 
 | |
5BU2
 
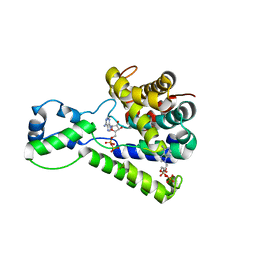 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, alpha-D-ribofuranose, ... | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTW
 
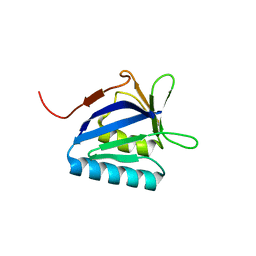 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
