1O3X
 
 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
7DDA
 
 | | Envelope protein VP37 a crystal structure from White Spot Syndrome Virus | | Descriptor: | Envelope protein, SULFATE ION | | Authors: | Somsoros, W, Sangawa, T, Takebe, K, Attarataya, J, Suzuki, M, Khunrae, P. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of the C-terminal domain of envelope protein VP37 from white spot syndrome virus reveals sulphate binding sites responsible for heparin binding.
J.Gen.Virol., 102, 2021
|
|
5GVO
 
 | | Solution NMR structure of a new lasso peptide sphaericin | | Descriptor: | Uncharacterized protein | | Authors: | Hemmi, H, Kodani, S, Inoue, Y, Suzuki, M, Dohra, H, Suzuki, T, Ohnishi-Kameyama, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-05 | | Method: | SOLUTION NMR | | Cite: | Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica
Eur.J.Org.Chem., 2017
|
|
3VQF
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | E3 ubiquitin-protein ligase LNX | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
3VK9
 
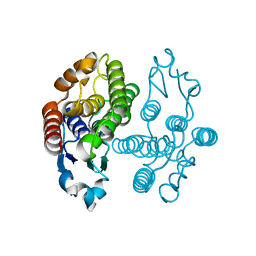 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
3VRP
 
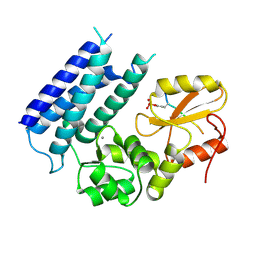 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRN
 
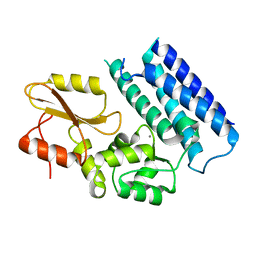 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3FI6
 
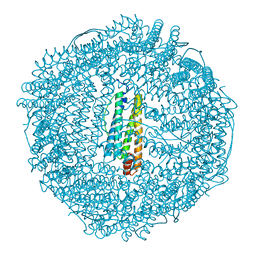 | | apo-H49AFr with high content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, PALLADIUM ION, ... | | Authors: | Abe, M, Ueno, T, Hirata, K, Suzuki, M, Abe, S, Shimizu, N, Yamaoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Process of Accumulation of Metal Ions on the Interior Surface of apo-Ferritin: Crystal Structures of a Series of apo-Ferritins Containing Variable Quantities of Pd(II) Ions
J.Am.Chem.Soc., 131, 2009
|
|
1GCC
 
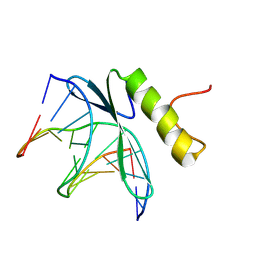 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | Authors: | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3GCC
 
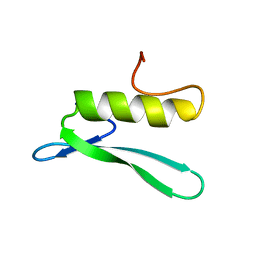 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
7CE4
 
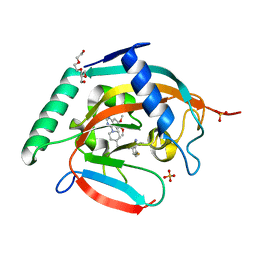 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
3VPQ
 
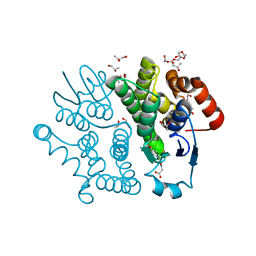 | | Crystal structure of Bombyx mori sigma-class glutathione transferase in complex with glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, Glutathione S-transferase sigma, ... | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A, Suzuki, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of a Bombyx mori sigma-class glutathione transferase exhibiting prostaglandin E synthase activity
Biochim.Biophys.Acta, 1830, 2013
|
|
3VUR
 
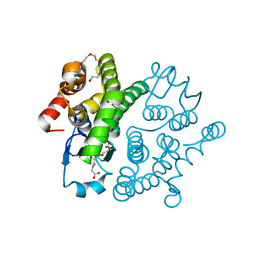 | |
2K7W
 
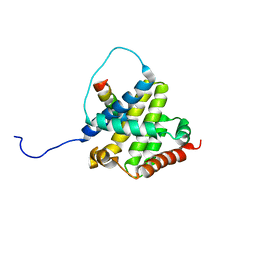 | | BAX Activation is Initiated at a Novel Interaction Site | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Gavathiotis, E, Suzuki, M, Davis, M.L, Pitter, K, Bird, G.H, Katz, S.G, Tu, H.C, Kim, H, Cheng, E.H, Tjandra, N, Walensky, L.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAX activation is initiated at a novel interaction site.
Nature, 455, 2008
|
|
5SW1
 
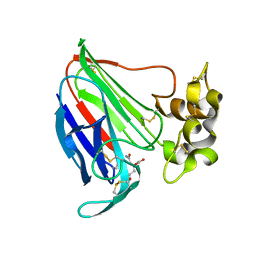 | | Thaumatin Structure at pH 6.0 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thaumatin Structure at pH 6.0
To Be Published
|
|
5SW2
 
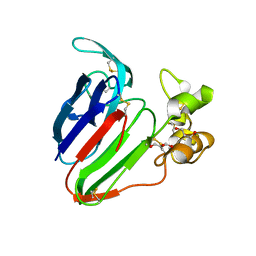 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
2HI7
 
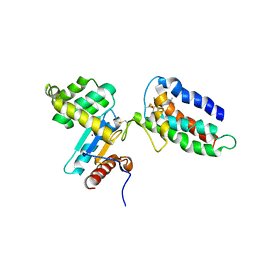 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
1HI3
 
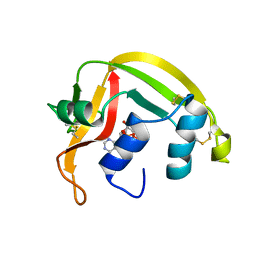 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI2
 
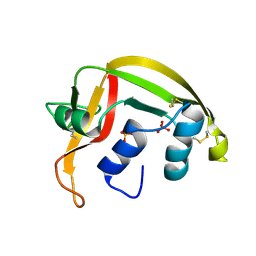 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
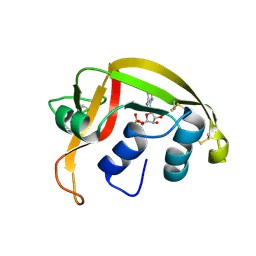 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
5SW0
 
 | |
4YOP
 
 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YSP
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSO
 
 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
