2YVX
 
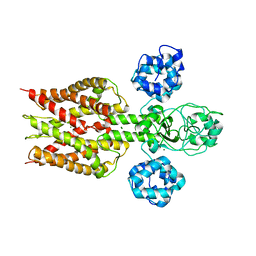 | | Crystal structure of magnesium transporter MgtE | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2YVZ
 
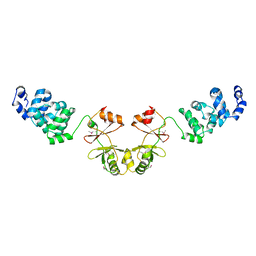 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+-free form | | Descriptor: | Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2YVY
 
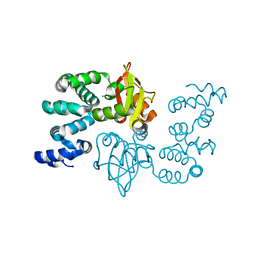 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+ bound form | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
4MKP
 
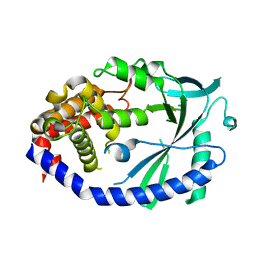 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
4N0H
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4N0I
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
7XJI
 
 | | Solabegron-activated dog beta3 adrenergic receptor | | Descriptor: | 3-[3-[2-[[(2~{S})-2-(3-chlorophenyl)-2-oxidanyl-ethyl]amino]ethylamino]phenyl]benzoic acid, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the beta 3 adrenergic receptor bound to solabegron and isoproterenol.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
4OO8
 
 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
4PDN
 
 | | Crystal structure of E. coli YfcM | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.
Nucleic Acids Res., 42, 2014
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
4U4V
 
 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | Descriptor: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
1UDS
 
 | | Crystal structure of the tRNA processing enzyme RNase PH R126A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UDN
 
 | | Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UDZ
 
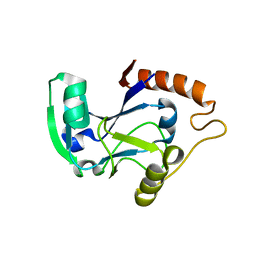 | | Isoleucyl-tRNA synthetase editing domain | | Descriptor: | Isoleucyl-tRNA synthetase | | Authors: | Fukunaga, R, Fukai, S, Ishitani, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine.
J.Biol.Chem., 279, 2004
|
|
1UDQ
 
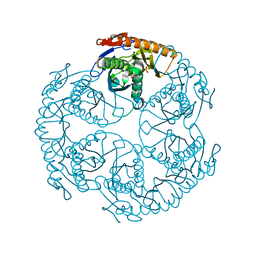 | | Crystal structure of the tRNA processing enzyme RNase PH T125A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UE0
 
 | | Isoleucyl-tRNA synthetase editing domain complexed with L-Valine | | Descriptor: | Isoleucyl-tRNA synthetase, VALINE | | Authors: | Fukunaga, R, Fukai, S, Ishitani, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine.
J.Biol.Chem., 279, 2004
|
|
1UDO
 
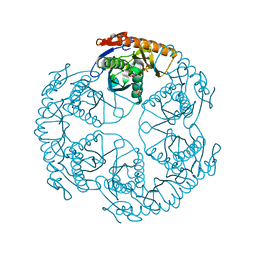 | | Crystal structure of the tRNA processing enzyme RNase PH R86A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UFA
 
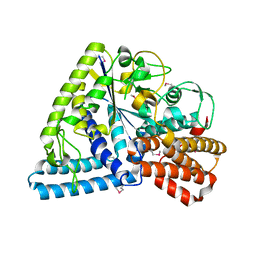 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQI
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
