2D0A
 
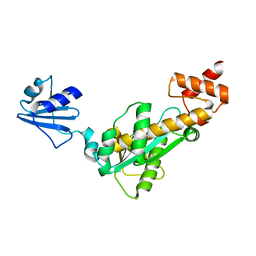 | | Crystal structure of Bst-RNase HIII | | Descriptor: | ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
3WQU
 
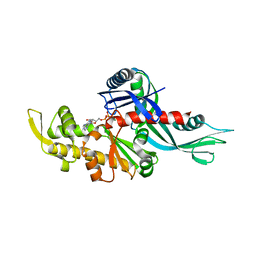 | | Staphylococcus aureus FtsA complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
3X2S
 
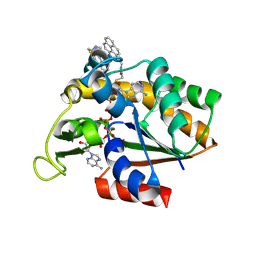 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
2DF5
 
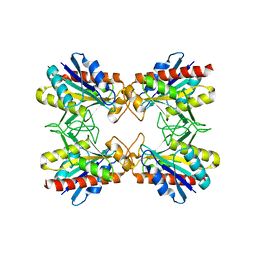 | | Crystal Structure of Pf-PCP(1-204)-C | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-02-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational contagion in a protein: structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DFH
 
 | | Crystal structure of Tk-RNase HII(1-212)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
3VGW
 
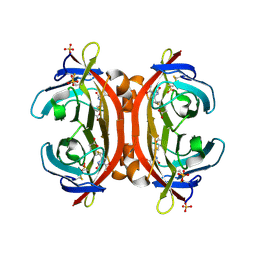 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
1FIY
 
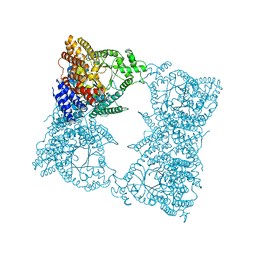 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3VND
 
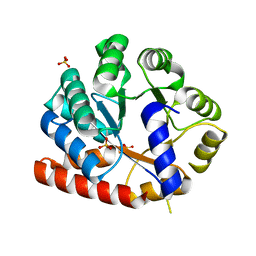 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
3VHI
 
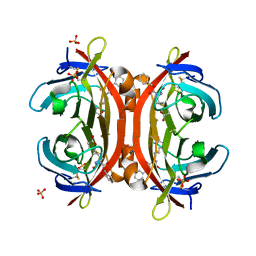 | | Crystal structure of monoZ-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{(3aS,4S,6aR)-1-[(benzyloxy)carbonyl]-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl}pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VI2
 
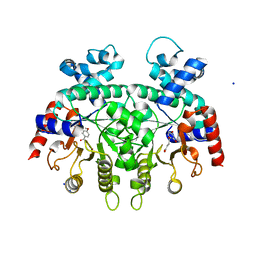 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
2E1P
 
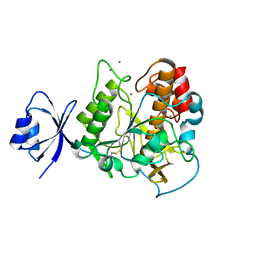 | | Crystal structure of pro-Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Saito, K, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of unautoprocessed precursor of subtilisin from a hyperthermophilic archaeon: evidence for Ca2+-induced folding
J.Biol.Chem., 282, 2007
|
|
2E4L
 
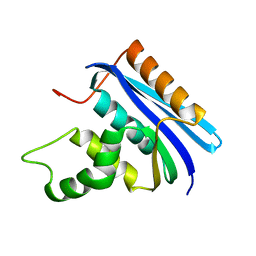 | | Thermodynamic and Structural Analysis of Thermolabile RNase HI from Shewanella oneidensis MR-1 | | Descriptor: | Ribonuclease HI | | Authors: | Tadokoro, T, You, D.J, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-12-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, thermodynamic, and mutational analyses of a psychrotrophic RNase HI.
Biochemistry, 46, 2007
|
|
3VHH
 
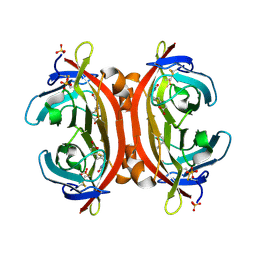 | | Crystal structure of DiMe-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1,3-dimethyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VHM
 
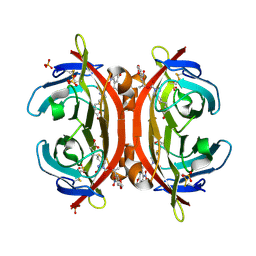 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3WQT
 
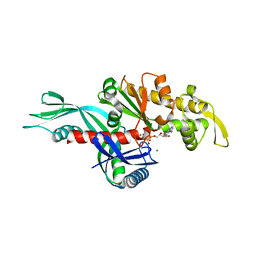 | | Staphylococcus aureus FtsA complexed with AMPPNP | | Descriptor: | CHLORIDE ION, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
3W6G
 
 | | Structure of peroxiredoxin from anaerobic hyperthermophilic archaeon Pyrococcus horikoshii | | Descriptor: | CITRATE ANION, Probable peroxiredoxin | | Authors: | Nakamura, T, Mori, A, Niiyama, M, Matsumura, H, Tokuyama, C, Morita, J, Uegaki, K, Inoue, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peroxiredoxin from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2DFI
 
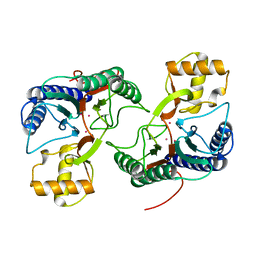 | | Crystal structure of Pf-MAP(1-292)-C | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DFE
 
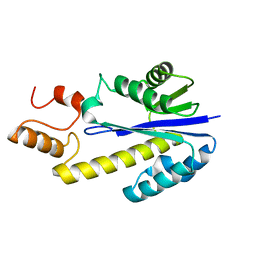 | | Crystal structure of Tk-RNase HII(1-200)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DFF
 
 | | Crystal structure of Tk-RNase HII(1-204)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
6K76
 
 | |
6KVQ
 
 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6KVP
 
 | | S. aureus FtsZ in complex with 3-(1-(5-bromo-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)ethoxy)-2,6-difluorobenzamide (compound 2) | | Descriptor: | 3-[(1R)-1-[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]ethoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6M4E
 
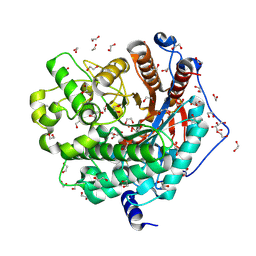 | | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M55
 
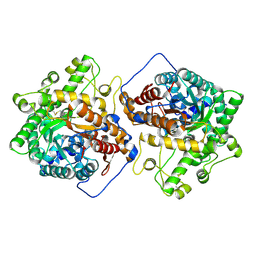 | | Crystal structure of the E496A mutant of HsBglA in complex with 4-galactosyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
6M4F
 
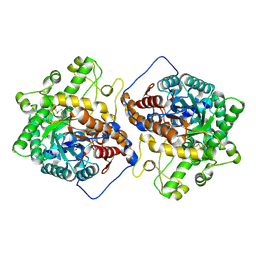 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
