5G1S
 
 | | Open conformation of Francisella tularensis ClpP at 1.7 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Diaz-Saez, L, Pankov, G, Hunter, W.N. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open and compressed conformations of Francisella tularensis ClpP.
Proteins, 85, 2017
|
|
2GBQ
 
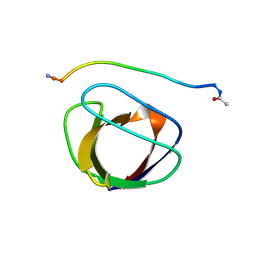 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
3DVX
 
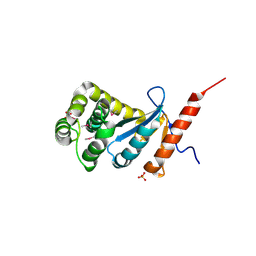 | | Crystal structure of reduced DsbA3 from Neisseria meningitidis | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Serre, L. | | Deposit date: | 2008-07-21 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
5B0M
 
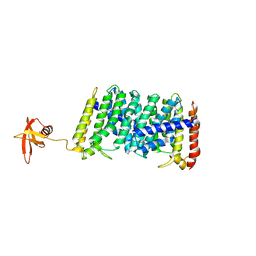 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-dodecyl maltoside | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
5GPR
 
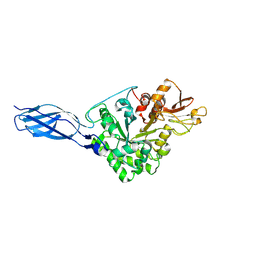 | | Crystal structure of chitinase-h from Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure, Catalysis, and Inhibition of OfChi-h, the Lepidoptera-exclusive Insect Chitinase.
J. Biol. Chem., 292, 2017
|
|
5BK6
 
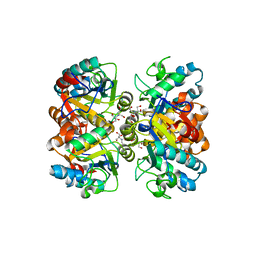 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-12 | | Release date: | 2018-02-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ANB
 
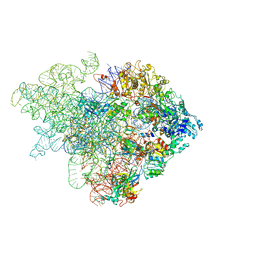 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AN5
 
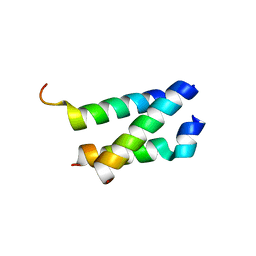 | | B. subtilis GpsB C-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, GLYCEROL | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
5GND
 
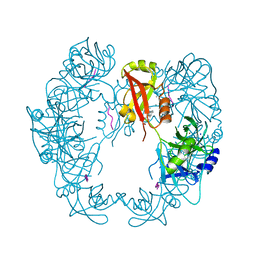 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
5B0I
 
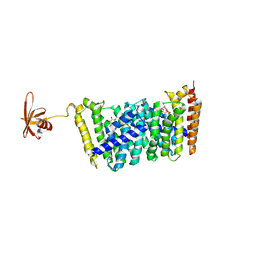 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-octyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, octyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5ISE
 
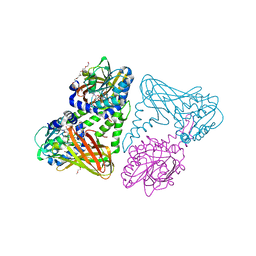 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0649 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BRM
 
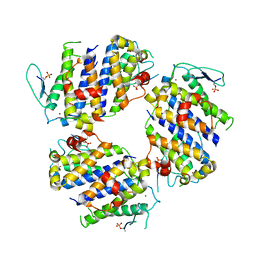 | |
5ALS
 
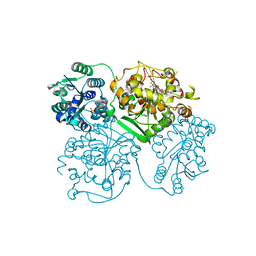 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, N-CYCLOPENTYL-2-[4-(TRIFLUOROMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE-7-SULFONAMIDE, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5IN3
 
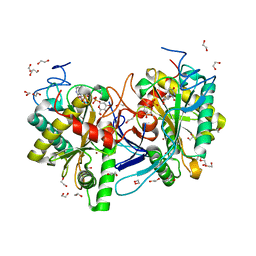 | | Crystal structure of glucose-1-phosphate bound nucleotidylated human galactose-1-phosphate uridylyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kopec, J, McCorvie, T, Tallant, C, Velupillai, S, Shrestha, L, Fitzpatrick, F, Patel, D, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis of classic galactosemia from the structure of human galactose 1-phosphate uridylyltransferase.
Hum.Mol.Genet., 25, 2016
|
|
5IO3
 
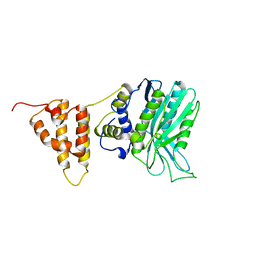 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5IRO
 
 | |
5IS7
 
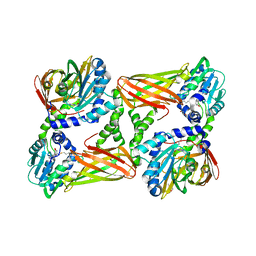 | | Crystal structure of mouse CARM1 in complex with decarboxylated SAH | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(3-aminopropyl)-5'-thioadenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with decarboxylated SAH
To Be Published
|
|
5ISI
 
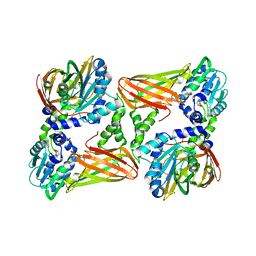 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine) | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 5'-amino-5'-deoxyadenosine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine)
To Be Published
|
|
5I0C
 
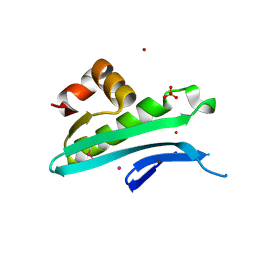 | | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12 | | Descriptor: | CADMIUM ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12
To Be Published
|
|
5BRR
 
 | | Michaelis complex of tPA-S195A:PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Crystal Structure of the Michaelis Complex between Tissue-type Plasminogen Activator and Plasminogen Activators Inhibitor-1
J.Biol.Chem., 290, 2015
|
|
5AR7
 
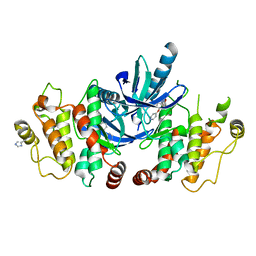 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biaryl Urea | | Descriptor: | 1-(5-TERT-BUTYL-1,2-OXAZOL-3-YL)-3-(4-PYRIDIN-4-YLOXYPHENYL)UREA, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
5AN8
 
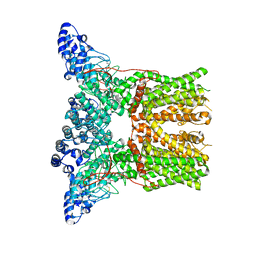 | | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | | Descriptor: | TRPV2 | | Authors: | Zubcevic, L, Herzik, M.A.J, Chung, B.C, Lander, G.C, Lee, S.Y. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-Electron Microscopy of the Trpv2 Ion Channel
Nat.Struct.Mol.Biol., 23, 2016
|
|
