7ELM
 
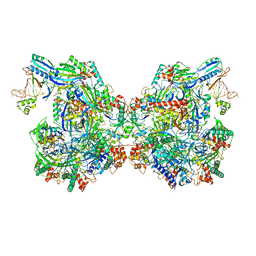 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
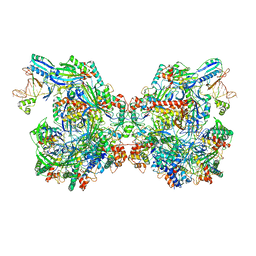 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
4AUJ
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to para hydroxypropargyl phenyl | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
7DTR
 
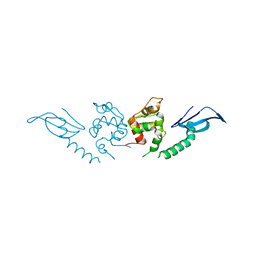 | | Structure of an AcrIF protein | | Descriptor: | AcrIF24 | | Authors: | Yue, F, Peipei, Y. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7F26
 
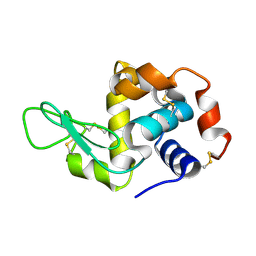 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Liang, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8VFX
 
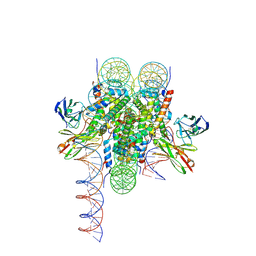 | | Cryo-EM structure of 186bp ALBN1 nucleosome aided by scFv | | Descriptor: | DNA (158-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
7XA7
 
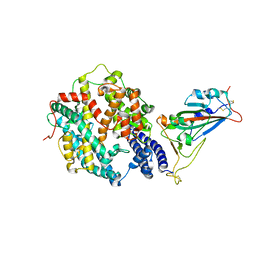 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
8WKU
 
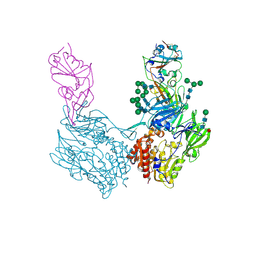 | | Complex structure of MjHKU4r-CoV-1 spike RBD bound to human CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, MjHKU4r-CoV-1 spike RBD, ... | | Authors: | Zhao, Z.N, Chai, Y, Gao, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for receptor recognition and broad host tropism for merbecovirus MjHKU4r-CoV-1.
Embo Rep., 25, 2024
|
|
7X8Q
 
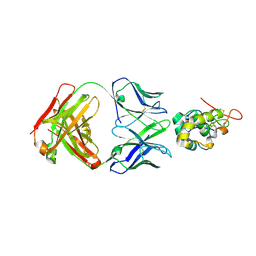 | | Frizzled-10 CRD in complex with F10_A9 Fab | | Descriptor: | Antibody F10_A9 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8T
 
 | | Frizzled 10 CRD in complex with hB9L9.3 Fab | | Descriptor: | Antibody hB9L9.3 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8P
 
 | | Frizzled 2 CRD in complex with pF7_A5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody pF7_A5 Fab, Heavy chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7QUO
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
4ATT
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl para methoxy phenyl | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
7F24
 
 | |
7F0T
 
 | |
7F23
 
 | |
7F1O
 
 | |
7F1Z
 
 | |
7XEM
 
 | | Cholesterol bound state of mTRPV2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEV
 
 | | Structure of mTRPV2_2-APB | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEO
 
 | | MbetaCD treated state of mTRPV2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XER
 
 | | Structure of mTRPV2_Q525T | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEU
 
 | | Structure of mTRPV2_E2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEW
 
 | | Structure of mTRPV2_Q525F | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
5ZAZ
 
 | |
