7K9M
 
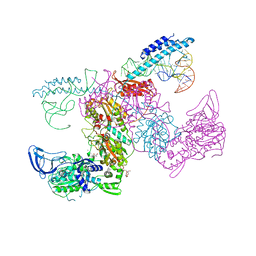 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
4ZO4
 
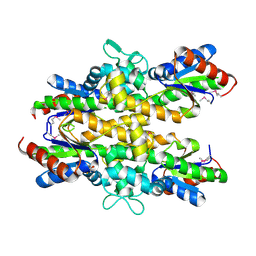 | | Dephospho-CoA kinase from Campylobacter jejuni. | | Descriptor: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | Authors: | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
1ZFJ
 
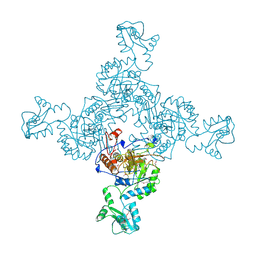 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
4ZPJ
 
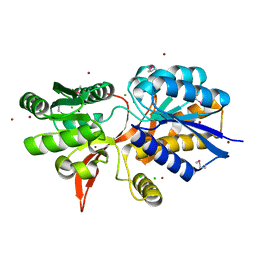 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | Authors: | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
3UK0
 
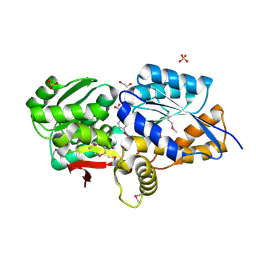 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3V7B
 
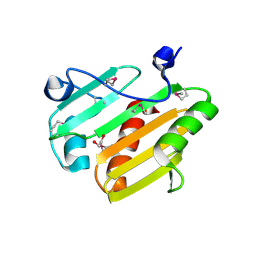 | | Dip2269 protein from corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein | | Authors: | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
3UO3
 
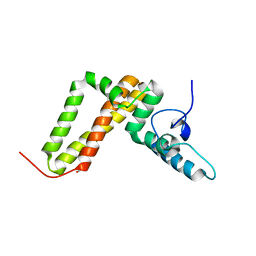 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | Descriptor: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3USB
 
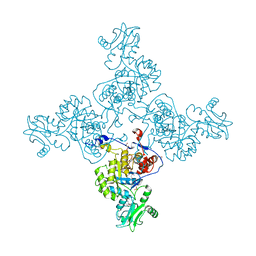 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4ZNM
 
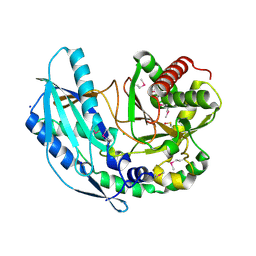 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | Descriptor: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZV9
 
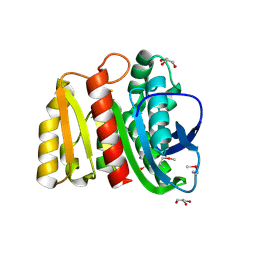 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-18 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
7KB3
 
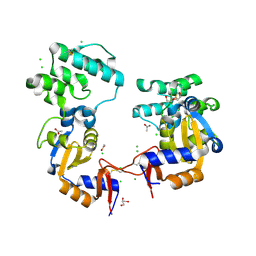 | | The structure of a sensor domain of a histidine kinase (VxrA) from Vibrio cholerae O1 biovar eltor str. N16961, 2nd form | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6NBK
 
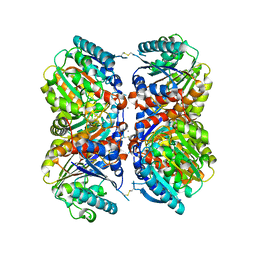 | | Crystal structure of Arginase from Bacillus cereus | | Descriptor: | Arginase, CALCIUM ION, MANGANESE (II) ION | | Authors: | Chang, C, Evdokimova, E, Mcchesney, M, Joachimiak, A, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Arginase from Bacillus cereus
To Be Published
|
|
7KB7
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
3S81
 
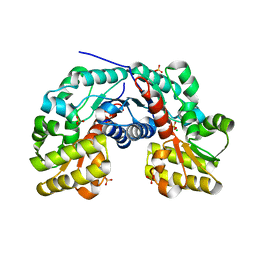 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
3S7Z
 
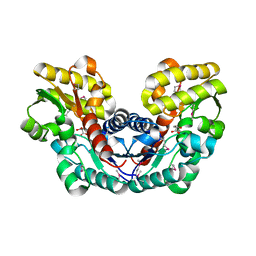 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate | | Descriptor: | MAGNESIUM ION, Putative aspartate racemase, SUCCINIC ACID, ... | | Authors: | Maltseva, N, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate.
To be Published
|
|
3SJR
 
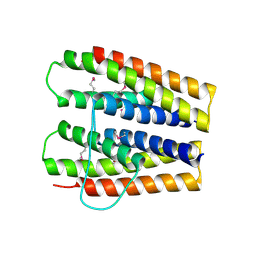 | |
3SHO
 
 | |
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
3S52
 
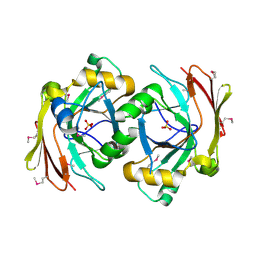 | | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, Putative fumarylacetoacetate hydrolase family protein, SULFATE ION | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92
TO BE PUBLISHED
|
|
3S83
 
 | | EAL domain of phosphodiesterase PdeA | | Descriptor: | GGDEF family protein, POTASSIUM ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of EAL domain from Caulobacter crescentus CB15
To be Published
|
|
3UGS
 
 | | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, Undecaprenyl pyrophosphate synthase | | Authors: | Nocek, B, Gu, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni
TO BE PUBLISHED
|
|
