7PB4
 
 | | Cenp-HIK 3-protein complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K | | Authors: | Bellini, D, Yatskevich, S, Muir, W.K, Barford, D. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7SQK
 
 | | Cryo-EM structure of the human augmin complex | | Descriptor: | HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 2, HAUS augmin-like complex subunit 3, ... | | Authors: | Gabel, C.A, Chang, L. | | Deposit date: | 2021-11-05 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Molecular architecture of the augmin complex.
Nat Commun, 13, 2022
|
|
1Q38
 
 | | Anastellin | | Descriptor: | Fibronectin | | Authors: | Briknarova, K, Akerman, M.E, Hoyt, D.W, Ruoslahti, E, Ely, K.R. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors
J.Mol.Biol., 332, 2003
|
|
7THK
 
 | |
7LLQ
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
7UOQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(5M)-5-{2-[(2-chloro-6-methylphenyl)methyl]-1,2,3,4-tetrahydroisoquinolin-6-yl}-4-(4,4-dimethylpiperidin-1-yl)-2-methylpyridin-3-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8867 Å) | | Cite: | Discovery and Preclinical Profiling of GSK3839919, a Potent HIV-1 Allosteric Integrase Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
5T2P
 
 | | Hepatitis B virus core protein Y132A mutant in complex with sulfamoylbenzamide (SBA_R01) | | Descriptor: | 4-fluoranyl-3-(4-oxidanylpiperidin-1-yl)sulfonyl-~{N}-[3,4,5-tris(fluoranyl)phenyl]benzamide, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
5TWM
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
3C6H
 
 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
8X61
 
 | | Cryo-EM structure of ATP-bound FtsE(E163Q)X | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX | | Authors: | Zhang, Z.Y, Chen, Y.T. | | Deposit date: | 2023-11-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
3CCC
 
 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(aminomethyl)-6-(2-chlorophenyl)-1-methyl-1H-benzimidazole-5-carbonitrile, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3C6A
 
 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
3CCB
 
 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 1-biphenyl-2-ylmethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CPE
 
 | | Crystal Structure of T4 gp17 | | Descriptor: | DNA packaging protein Gp17, PHOSPHATE ION, SODIUM ION | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-03-31 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces
Cell(Cambridge,Mass.), 135, 2008
|
|
3E7J
 
 | | HeparinaseII H202A/Y257A double mutant complexed with a heparan sulfate tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Heparinase II protein, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2008-08-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate.
J.Biol.Chem., 285, 2010
|
|
7PKN
 
 | | Structure of the human CCAN deltaCT complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Muir, K.W, Yatskevich, S, Bellini, D, Barford, D. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7PII
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (122-MER), DNA (123-MER), ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Barford, D. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7QRI
 
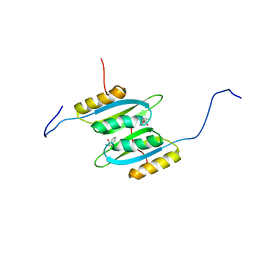 | | Regulatory domain dimer of tryptophan hydroxylase 2 in complex with L-Phe | | Descriptor: | PHENYLALANINE, Tryptophan 5-hydroxylase 2 | | Authors: | Vedel, I.M, Prestel, A, Harris, P, Peters, G.H.J, Kragelund, B.B. | | Deposit date: | 2022-01-11 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human tryptophan hydroxylase 2 reveals that L-Phe is superior to L-Trp as the regulatory domain ligand.
Structure, 31, 2023
|
|
7RJ5
 
 | | The structure of BAM in complex with EspP at 7 Angstrom resolution | | Descriptor: | Maltodextrin-binding protein,Autotransporter outer membrane beta-barrel domain-containing protein chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI5
 
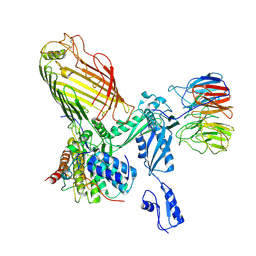 | | Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI6
 
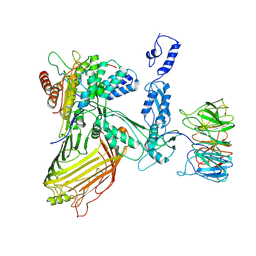 | |
7RI4
 
 | | Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate | | Descriptor: | EspPbeta9-12, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI7
 
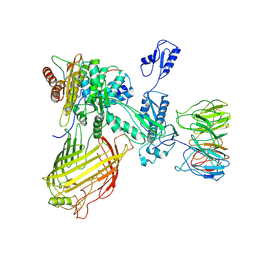 | | The structure of BAM in MSP1D1 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
