6BIA
 
 | | Crystal structure of Ps i-CgsB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6OPO
 
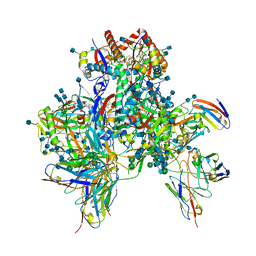 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPN
 
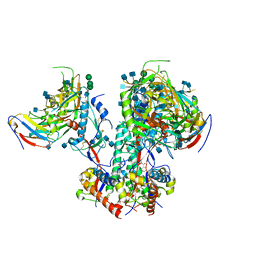 | | CD4- and 17-bound HIV-1 Env B41 SOSIP in complex with small molecule GO35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPP
 
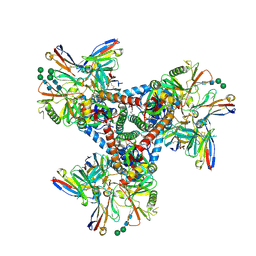 | | Asymmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPQ
 
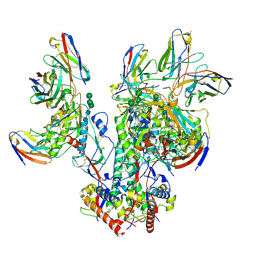 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6X5C
 
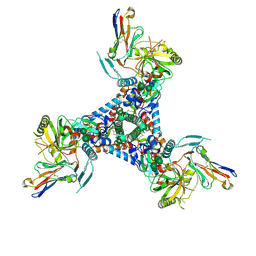 | | Asymmetric model of CD4-bound B41 HIV-1 Env SOSIP in complex with small molecule GO52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2020-05-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6X5B
 
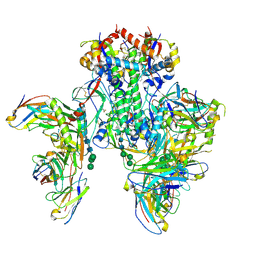 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with small molecule GO52 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2020-05-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
2WJM
 
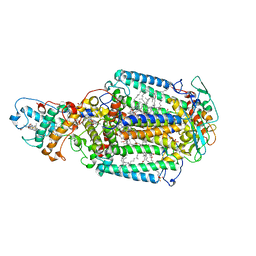 | | Lipidic sponge phase crystal structure of the photosynthetic reaction centre from Blastochloris viridis (low dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Woehri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
4BQT
 
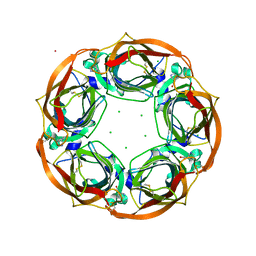 | | Aplysia californica AChBP in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Rucktooa, P, Haseler, C.A, vanElke, R, Smit, A.B, Gallagher, T, Sixma, T.K. | | Deposit date: | 2013-06-02 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Characterization of Binding Mode of Smoking Cessation Drugs to Nicotinic Acetylcholine Receptors Through Study of Ligand Complexes with Acetylcholine-Binding Protein.
J.Biol.Chem., 287, 2012
|
|
4C23
 
 | | L-fuculose kinase | | Descriptor: | 1,2-ETHANEDIOL, L-FUCULOSE KINASE FUCK | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4D6C
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6D
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4C21
 
 | | L-Fucose Isomerase In Complex With Fucitol | | Descriptor: | 1,2-ETHANEDIOL, FUCITOL, L-FUCOSE ISOMERASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4BMQ
 
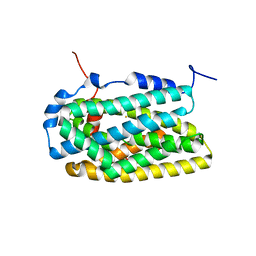 | | Crystal Structure of Ribonucleotide Reductase apo-NrdF from Bacillus cereus (space group C2) | | Descriptor: | FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Tomter, A.B, Hersleth, H.-P, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4BQ3
 
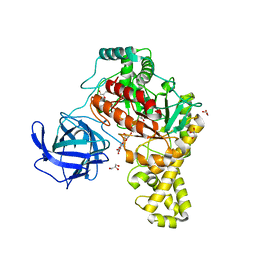 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4BMU
 
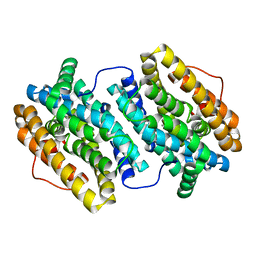 | | Crystal Structure of Ribonucleotide Reductase di-manganese(II) NrdF from Bacillus cereus | | Descriptor: | MANGANESE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Hersleth, H.-P, Tomter, A.B, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4C24
 
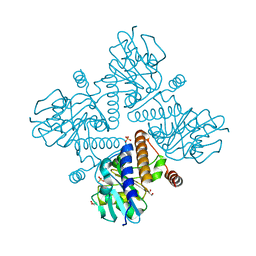 | | L-fuculose 1-phosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-FUCULOSE PHOSPHATE ALDOLASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4BMT
 
 | | Crystal Structure of Ribonucleotide Reductase di-iron NrdF from Bacillus cereus | | Descriptor: | FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Hersleth, H.-P, Tomter, A.B, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4D6J
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6G
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D71
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6E
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4DMI
 
 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences (CASP) | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, SODIUM ION | | Authors: | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-02-07 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences
To be Published
|
|
4D6F
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
