2A8V
 
 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
2AF5
 
 | | 2.5A X-ray Structure of Engineered OspA protein | | Descriptor: | Engineered Outer Surface Protein A (OspA) with the inserted two beta-hairpins | | Authors: | Makabe, K, Mcelheny, D, Tereshko, V, Hilyard, A, Koide, A, Koide, S. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AFI
 
 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
4CSV
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl bound to Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, SRC-ABL TYROSINE KINASE ANCESTOR | | Authors: | Kutter, S, Wilson, C, Cruz, L, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Kern, D. | | Deposit date: | 2014-03-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
2AB5
 
 | | bI3 LAGLIDADG Maturase | | Descriptor: | SULFATE ION, mRNA maturase | | Authors: | Longo, A, Leonard, C.W, Bassi, G.S, Berndt, D, Krahn, J.M, Hall, T.M, Weeks, K.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2ADY
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex IV) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1ZYH
 
 | | Structure of a Supercoiling Responsive DNA site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*GP*GP*GP*TP*TP*G)-3', 5'-D(*CP*AP*AP*CP*CP*CP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
2AE9
 
 | | Solution Structure of the theta subunit of DNA polymerase III from E. coli | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Mueller, G.A, Kirby, T.W, Derose, E.F, Li, D, Schaaper, R.M, London, R.E. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
J.Bacteriol., 187, 2005
|
|
1ZF1
 
 | | CCC A-DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFA
 
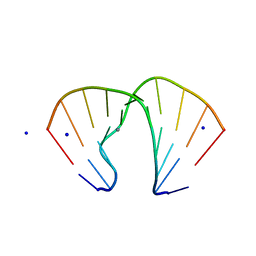 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z6I
 
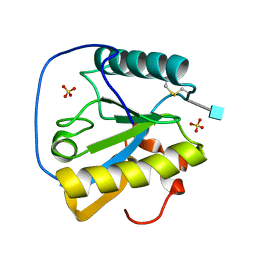 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZK7
 
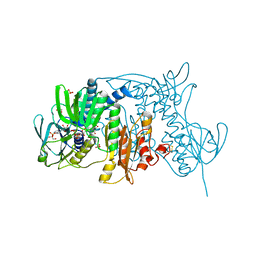 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
1ZPD
 
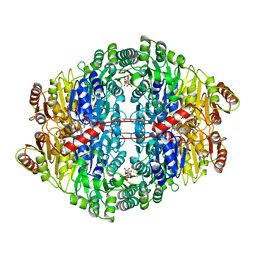 | | PYRUVATE DECARBOXYLASE FROM ZYMOMONAS MOBILIS | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MONO-{4-[(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-AMINO]-2-HYDROXY-3-MERCAPTO-PENT-3-ENYL-PHOSPHONO} ESTER, ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1998-04-17 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
J.Biol.Chem., 273, 1998
|
|
1ZF8
 
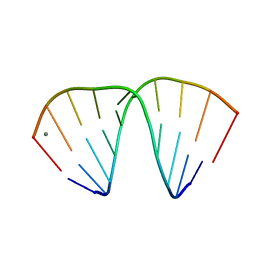 | | GGT Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*GP*GP*TP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z1G
 
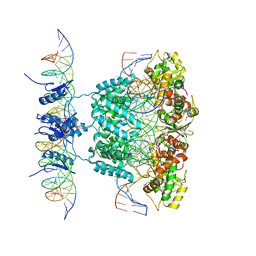 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | Descriptor: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1Z9B
 
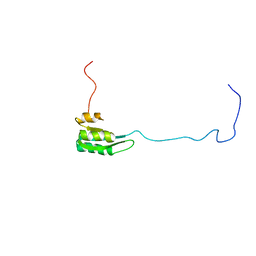 | | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tomaselli, S, Bernard, C, Spurio, R, Picone, D, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2005-04-01 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2
Protein Sci., 14, 2005
|
|
2A2X
 
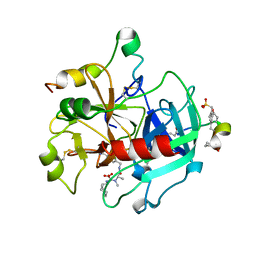 | | Orally Active Thrombin Inhibitors in Complex with Thrombin Inh12 | | Descriptor: | N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({6-[AMINO(IMINO)METHYL]PYRIDIN-3-YL}METHYL)-N~2~-METHYL-L-ALANINAMIDE, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Lange, U.E.W, Baucke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Orally active thrombin inhibitors. Part 2: optimization of the P2-moiety
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2A41
 
 | | Ternary complex of the WH2 Domain of WIP with Actin-DNAse I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-06-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4CZV
 
 | | Structure of the Neurospora crassa Pan2 WD40 domain | | Descriptor: | CHLORIDE ION, GLYCEROL, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN2 | | Authors: | Peter, D, Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Asymmetric Pan3 Dimer Recruits a Single Pan2 Exonuclease to Mediate Mrna Deadenylation and Decay.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2A8Z
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2005-07-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase.
J.Mol.Biol., 354, 2005
|
|
2ABA
 
 | | Structure of reduced PETN reductase in complex with progesterone | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPROPYL ALCOHOL, PROGESTERONE, ... | | Authors: | Khan, H, Barna, T, Bruce, N.C, Munro, A.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2005-07-15 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Proton transfer in the oxidative half-reaction of pentaerythritol tetranitrate reductase
Febs J., 272, 2005
|
|
2AC0
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2A1T
 
 | | Structure of the human MCAD:ETF E165betaA complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, Van Thiel, A, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stabilization of Non-productive Conformations Underpins Rapid Electron Transfer to Electron-transferring Flavoprotein
J.Biol.Chem., 280, 2005
|
|
2A5H
 
 | | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | | Descriptor: | IRON/SULFUR CLUSTER, L-lysine 2,3-aminomutase, LYSINE, ... | | Authors: | Lepore, B.W, Ruzicka, F.J, Frey, P.A, Ringe, D. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A92
 
 | | Crystal structure of lactate dehydrogenase from Plasmodium vivax: complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
