7MJ8
 
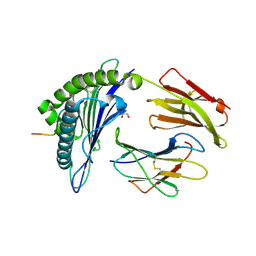 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7LZ7
 
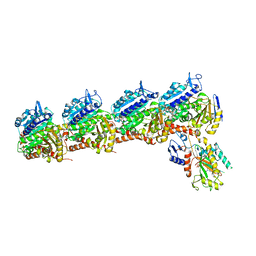 | | Tubulin-RB3_SLD-TTL in complex with compound 5k | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7LZ8
 
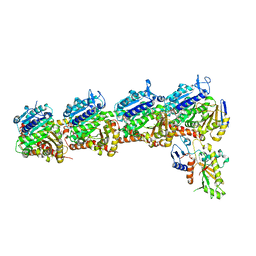 | | Tubulin-RB3_SLD-TTL in complex with compound 5t | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)pyrido[3,2-d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
5F8S
 
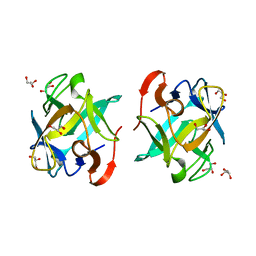 | | Crystal structure of a Crenomytilus grayanus lectin | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
5FAD
 
 | | SAH complex with aKMT from the hyperthermophilic archaeon Sulfolobus islandicus | | Descriptor: | MAGNESIUM ION, Ribosomal protein L11 methyltransferase, putative, ... | | Authors: | Ouyang, S. | | Deposit date: | 2015-12-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism
Mol.Cell Proteomics, 15, 2016
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
6JEH
 
 | |
7NAM
 
 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5HI4
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (9'S,17'R)-6'-chloro-N-methyl-9'-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-10',19'-dioxo-2'-oxa-11',18'-diazaspiro[cyclopentane-1,21'-tetracyclo[20.2.2.2~12,15~.1~3,7~]nonacosane]-1'(24'),3'(29'),4',6',12',14',22',25',27'-nonaene-17'-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 FAB light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
8X2Z
 
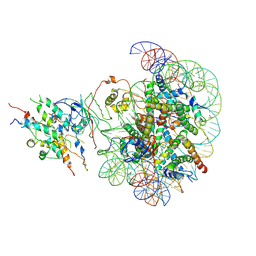 | | The class2 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X32
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome-H4KQ Complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X30
 
 | | Structure of piccolo NuA4 and H2A.Z nucleosome 2:1 complex | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2Y
 
 | | The class1 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2X
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex at pre-H4-acetylation state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X31
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4LPI
 
 | |
4NNH
 
 | |
4NMX
 
 | |
4NNI
 
 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
4NNG
 
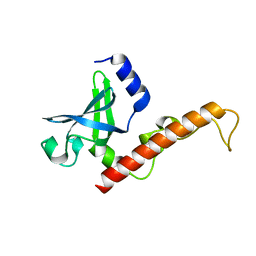 | |
4NNK
 
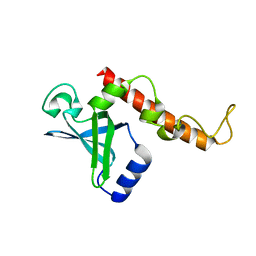 | |
7XB4
 
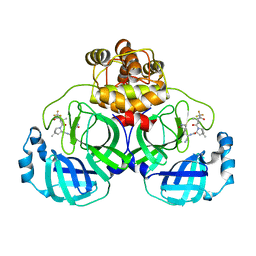 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7V67
 
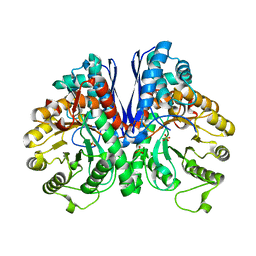 | |
