6B8O
 
 | | WT Ig-like V Domain with Phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Wang, Z. | | Deposit date: | 2017-10-09 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
8JNJ
 
 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8JNI
 
 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
6LJJ
 
 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LAU
 
 | | the wildtype SAM-VI riboswitch bound to SAH | | Descriptor: | CESIUM ION, GUANOSINE-5'-TRIPHOSPHATE, RNA (54-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LBS
 
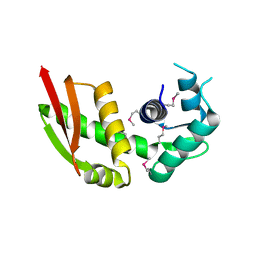 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7CV9
 
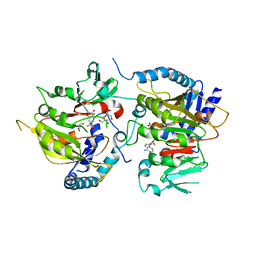 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV7
 
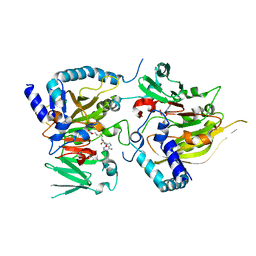 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
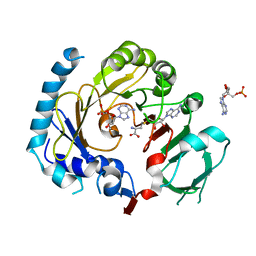 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV8
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, SINEFUNGIN | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7EKI
 
 | | human alpha 7 nicotinic acetylcholine receptor in apo-form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKP
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKT
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 and PNU-120596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
8XMS
 
 | |
8YHZ
 
 | | The co-crystal structure of the Fab fragment of Ab-1080 with NaV1.7 VSDII peptide | | Descriptor: | Heavy chain of 1080 Fab, Light chain of 1080 Fab, Sodium channel protein type 9 subunit alpha | | Authors: | Du, J, Zhang, Y, Zhu, R, Ding, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Intra-channel bi-epitopic crosslinking unleashes ultrapotent antibodies targeting Na V 1.7 for pain alleviation.
Cell Rep Med, 5, 2024
|
|
9AUU
 
 | | Hsp90 NTD in complex with compound 6 | | Descriptor: | 3-{[(3R)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}-6-fluoro-2-hydroxybenzaldehyde, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Weaver, J.M, Craven, G.B, Taunton, J. | | Deposit date: | 2024-02-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminomethyl Salicylaldehydes Lock onto a Surface Lysine by Forming an Extended Intramolecular Hydrogen Bond Network.
J.Am.Chem.Soc., 146, 2024
|
|
8Z2O
 
 | | Crystal structure of 5-N-alpha-glycinylthymidine (N-alpha-GlyT) FAD-dependent lyase gp47/NGTO from Pseudomonads phage PaMx11 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent lyase, ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z2M
 
 | |
8Z79
 
 | |
8Z2N
 
 | | Crystal structure of 5-phosphomethyl-2'-deoxyuridine (5-PmdU) glycinyltransferase gp46/PUGT from Pseudomonads phage PaMx11 in complex with dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*GP*TP*CP*GP*AP*CP*GP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*GP*TP*CP*GP*AP*CP*TP*A)-3'), ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8XKS
 
 | | The cryo-EM structure of Orf2971-FtsHi motor complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 4Fe-4S ferredoxin-type domain-containing protein, ... | | Authors: | Wang, N, Li, M. | | Deposit date: | 2023-12-24 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture of the ATP-driven motor for protein import into chloroplasts.
Mol Plant, 17, 2024
|
|
8X5F
 
 | | human XPR1 in complex with InsP6 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
8X5B
 
 | | Cryo-EM structures of human XPR1 in closed states | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, PHOSPHATE ION, ... | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
8X5E
 
 | | Cryo-EM structure of human XPR1 in open state | | Descriptor: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
