5AO4
 
 | |
1OZ1
 
 | | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR | | Descriptor: | 3-(4-FLUOROPHENYL)-2-PYRIDIN-4-YL-1H-PYRROLO[3,2-B]PYRIDIN-1-OL, Mitogen-activated protein kinase 14 | | Authors: | Lovejoy, B, Villasenor, A, Browner, M, Dunten, P. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of 4-azaindoles as inhibitors of p38 MAP kinase.
J.Med.Chem., 46, 2003
|
|
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
4X9D
 
 | | High-resolution structure of Hfq from Methanococcus jannaschii in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, E.Y, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
1VRM
 
 | |
6RHQ
 
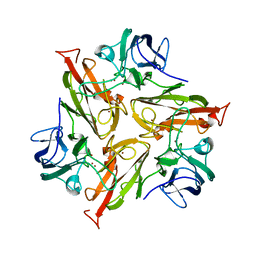 | | Crystal Structure of Two-Domain Laccase mutant I170A from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6RH9
 
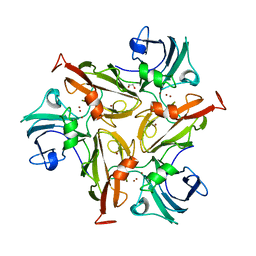 | | Crystal Structure of Two-Domain Laccase mutant I170F from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6IHJ
 
 | |
6S0O
 
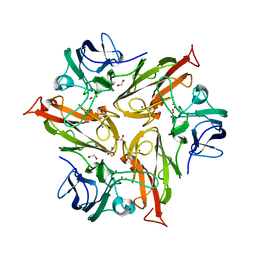 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
4X9C
 
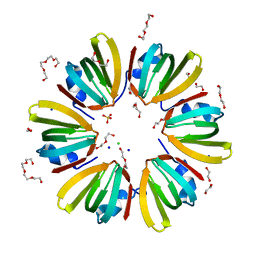 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
1QD7
 
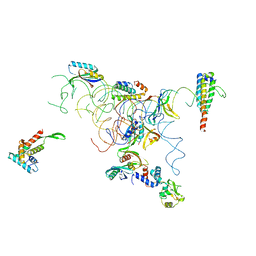 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1ZX8
 
 | |
5DY9
 
 | | Y68T Hfq from Methanococcus jannaschii in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nikulin, A.D, Mikhailina, A.O, Lekontseva, N.V, Balobanov, V.A, Nikonova, E.Y, Tishchenko, S.V. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
1ZKG
 
 | |
4PPD
 
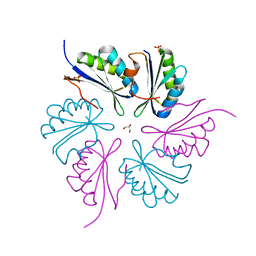 | | PduA K26A, crystal form 2 | | Descriptor: | GLYCEROL, Propanediol utilization protein PduA, SULFATE ION | | Authors: | McNamara, D.E, Sawaya, M.R, Bobik, T.A, Yeates, T.O. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the pdu microcompartment.
J.Mol.Biol., 426, 2014
|
|
4O2P
 
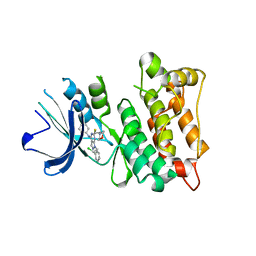 | | Kinase domain of cSrc in complex with a substituted pyrazolopyrimidine | | Descriptor: | 1-[(2R)-2-chloro-2-phenylethyl]-6-{[2-(morpholin-4-yl)ethyl]sulfanyl}-N-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Richters, A, Rauh, D. | | Deposit date: | 2013-12-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining X-ray Crystallography and Molecular Modeling toward the Optimization of Pyrazolo[3,4-d]pyrimidines as Potent c-Src Inhibitors Active in Vivo against Neuroblastoma.
J.Med.Chem., 58, 2015
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
1VR0
 
 | |
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
1VR8
 
 | |
1UV0
 
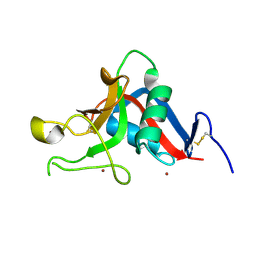 | | Pancreatitis-associated protein 1 from human | | Descriptor: | PANCREATITIS-ASSOCIATED PROTEIN 1, ZINC ION | | Authors: | Abergel, C, Shepard, W, Christal, L. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallization and preliminary crystallographic study of HIP/PAP, a human C-lectin overexpressed in primary liver cancers.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1VQ3
 
 | |
8P9V
 
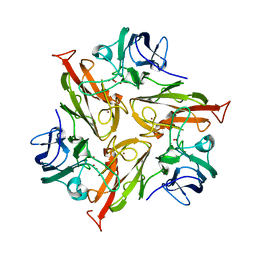 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H/D268N from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, PEROXIDE ION, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
8P9U
 
 | | Crystal Structure of Two-Domain Laccase mutant M199A/D268N from Streptomyces griseoflavus | | Descriptor: | CALCIUM ION, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
2K7M
 
 | | Structure of the Connexin40 Carboxyl terminal Domain | | Descriptor: | Gap junction alpha-5 protein | | Authors: | Bouvier, D, Spagnol, G, Kieken, F, Vitrac, H, Kellezi, A, Forge, V. | | Deposit date: | 2008-08-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Characterization of the structure and intermolecular interactions between the connexin40 and connexin43 carboxyl-terminal and cytoplasmic loop domains.
J.Biol.Chem., 284, 2009
|
|
