4ICK
 
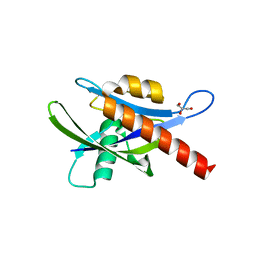 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4IJX
 
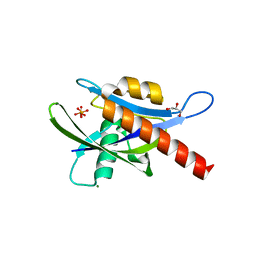 | |
4J3D
 
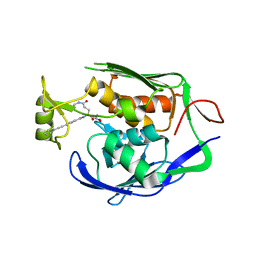 | | Pseudomonas aeruginosa LpxC in complex with a hydroxamate inhibitor | | Descriptor: | N~1~-hydroxy-N~5~-(3-hydroxypropyl)-N~2~-[4-(phenylethynyl)benzoyl]-L-glutamamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Lahiri, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the UDP pocket of LpxC through amino acid analogs.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | Descriptor: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
7VQ3
 
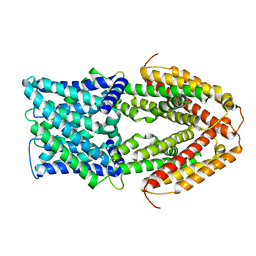 | |
7VQ5
 
 | |
7VQ4
 
 | |
7VQ7
 
 | | The Al-bound AtALMT1 structure at pH 5 (ALMT1Al/pH5) | | Descriptor: | ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J.Q. | | Deposit date: | 2021-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
2NAX
 
 | | Structure of CCHC zinc finger domain of Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3'-end processing.
RNA, 23, 2017
|
|
4QM0
 
 | | Crystal structure of RORc in complex with a tertiary sulfonamide inverse agonist | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-methylpropyl)-N-({5-[4-(methylsulfonyl)phenyl]thiophen-2-yl}methyl)-1-phenylmethanesulfonamide, Nuclear receptor ROR-gamma | | Authors: | Boenig, G, Hymowitz, S.G, Wang, W. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Reduction in lipophilicity improved the solubility, plasma-protein binding, and permeability of tertiary sulfonamide RORc inverse agonists.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INI
 
 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6ING
 
 | |
6INH
 
 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
7C7P
 
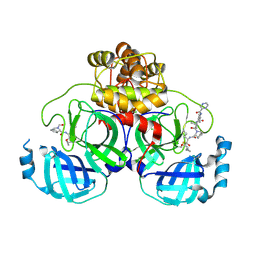 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6Y1Y
 
 | |
8X83
 
 | |
8X82
 
 | |
8X84
 
 | |
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
2UZQ
 
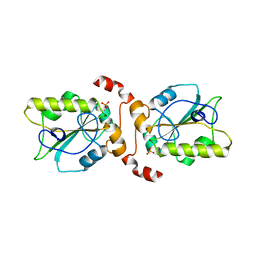 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
6L3R
 
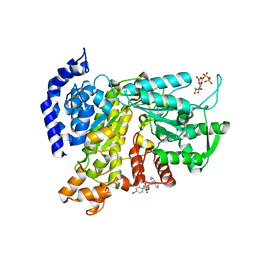 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | Descriptor: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
6L7L
 
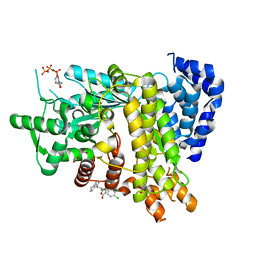 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide | | Descriptor: | 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
