3UXY
 
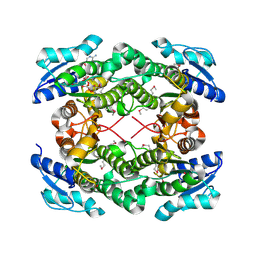 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
7D99
 
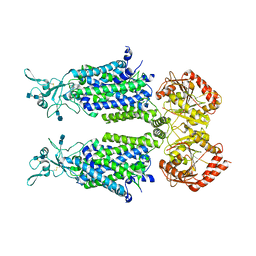 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
8SW4
 
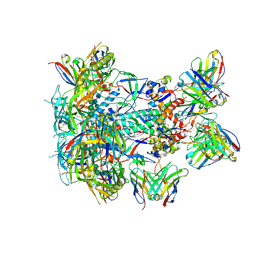 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site.
Sci Immunol, 9, 2024
|
|
7D8Z
 
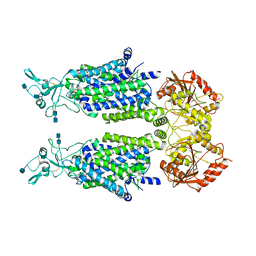 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D90
 
 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3K6G
 
 | | Crystal structure of Rap1 and TRF2 complex | | Descriptor: | Telomeric repeat-binding factor 2, Telomeric repeat-binding factor 2-interacting protein 1 | | Authors: | Chen, Y, Rai, R, Yang, Y.T, Zheng, H, Chang, S, Lei, M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4ZW1
 
 | | Crystal structure of hBRD4 in complex with BL-BI06 reveals a novel synthesized inhibitor that induces Beclin1-independent/ATG5-dependent autophagic cell death in breast cancer | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-hydroxy-3,5-dimethylphenyl)-7-methyl-5,6,7,8-tetrahydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Liu, B, Zhang, S, Guo, M, Tian, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of hBRD4 in complex with BL-BI06 reveals a novel synthesized inhibitor that induces Beclin1-independent/ATG5-dependent autophagic cell death in breast cancer
To Be Published
|
|
6Q84
 
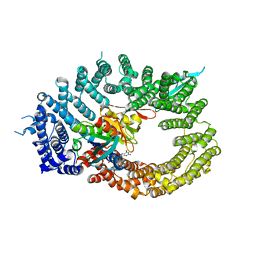 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
3UOE
 
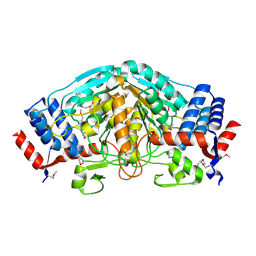 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
7QR3
 
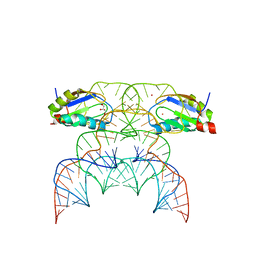 | | Chimpanzee CPEB3 HDV-like ribozyme | | Descriptor: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
7Q6L
 
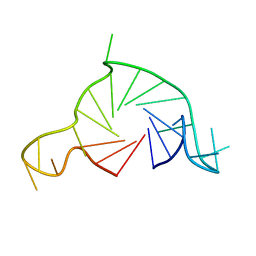 | |
8TOO
 
 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNN
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
3V5N
 
 | | The crystal structure of oxidoreductase from Sinorhizobium meliloti | | Descriptor: | Oxidoreductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-16 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | The crystal structure of oxidoreductase from Sinorhizobium meliloti
To be Published
|
|
7QR4
 
 | | human CPEB3 HDV-like ribozyme | | Descriptor: | RNA CPEB3 ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
3TG9
 
 | | The crystal structure of penicillin binding protein from Bacillus halodurans | | Descriptor: | Penicillin-binding protein | | Authors: | Zhang, Z, Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of penicillin binding protein from Bacillus halodurans
To be Published
|
|
6Q83
 
 | | Crystal structure of the biportin Pdr6 in complex with UBC9 | | Descriptor: | Importin beta-like protein KAP122, UBC9 | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6Q82
 
 | | Crystal structure of the biportin Pdr6 in complex with RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin beta-like protein KAP122, ... | | Authors: | Aksu, M, Vera-Rodriguez, A, Trakhanov, S, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6QGM
 
 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
7S16
 
 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S22
 
 | | Crystal structure of alpha-COP-WD40 domain | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S23
 
 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7V3F
 
 | | DENV2_NGC_Fab_C10 28degree (1Fab:3E) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
