1PDH
 
 | |
7ZNZ
 
 | | Crystal structure of unliganded form of FucOB, a GH95 family alpha-1,2-fucosidase from Akkermansia muciniphila | | Descriptor: | FucOB, a GH95 family alpha-1,2-fucosidase, GLYCEROL | | Authors: | Anso, I, Cifuente, J.O, Trastoy, B, Guerin, M.E. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Turning universal O into rare Bombay type blood.
Nat Commun, 14, 2023
|
|
8QOE
 
 | | Inward-facing conformation of the ABC transporter BmrA | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Valimehr, S, Hanssen, E, Orelle, C, Jault, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
1CC4
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1BGN
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 269 REPLACED BY THR (R269T), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
6ASP
 
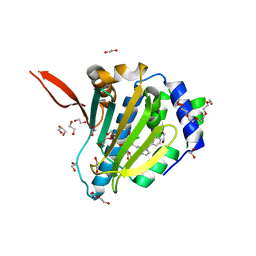 | | Structure of Grp94 with methyl 3-chloro-2-(2-(1-(2-ethoxybenzyl)-1 H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, a Grp94-selective inhibitor and promising therapeutic lead for treating myocilin-associated glaucoma | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, GLYCEROL, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
1CC6
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1PBF
 
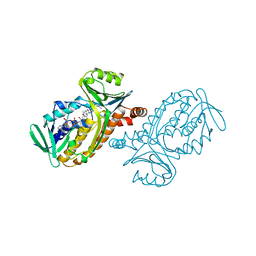 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBD
 
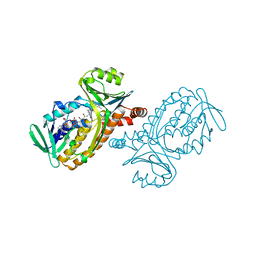 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
8B55
 
 | | Human ADGRG4 PTX-like domain | | Descriptor: | Adhesion G-protein coupled receptor G4, MAGNESIUM ION | | Authors: | Kieslich, B, Straeter, N. | | Deposit date: | 2022-09-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J.Biol.Chem., 299, 2023
|
|
7APK
 
 | | Structure of the human THO - UAP56 complex | | Descriptor: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | Authors: | Hohmann, U, Puehringer, T, Plaschka, C. | | Deposit date: | 2020-10-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human core transcription-export complex reveals a hub for multivalent interactions.
Elife, 9, 2020
|
|
8T2I
 
 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
7WGA
 
 | |
8CHB
 
 | | Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Hanssen, E, Valimehr, S, Orelle, C, Jault, J.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
1CJ3
 
 | | MUTANT TYR38GLU OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ4
 
 | | MUTANT Q34T OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ2
 
 | | MUTANT GLN34ARG OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
6ASQ
 
 | | Structure of Grp94 bound to methyl 2-[2-(2-benzylpyridin-3-yl)ethyl]-3-chloro-4,6-dihydroxybenzoate, a pan-Hsp90 inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
6BI2
 
 | |
6BHZ
 
 | | Trastuzumab Fab D185A (Light Chain) Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab Anti-HER2 Fab Heavy Chain, Trastuzumab Anti-HER2 Fab Light Chain D185A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6F2C
 
 | | Methylglyoxal synthase MgsA from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Methylglyoxal synthase | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-11-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the regulatory interaction of the methylglyoxal synthase MgsA with the carbon flux regulator Crh inBacillus subtilis.
J. Biol. Chem., 293, 2018
|
|
6BI0
 
 | | Trastuzumab Fab N158A, D185A, K190A (Light Chain) Triple Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab anti-HER2 Fab Heavy Chain, Trastuzumab anti-HER2 Fab Light Chain | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
3QUP
 
 | |
6I6Z
 
 | |
4Z8L
 
 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
