7VSK
 
 | | Crystal structure of E.coli pseudouridine kinase PsuK complexed with pseudouridine. | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, PfkB domain protein | | Authors: | Li, K.J, Li, X.J, Wu, B.X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.
Front Microbiol, 13, 2022
|
|
7W93
 
 | | Crystal structure of E.coli pseudouridine kinase PsuK complexed with N1-methyl-pseudouridine | | Descriptor: | 5-[(2S,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-pyrimidine-2,4-dione, POTASSIUM ION, PfkB domain protein | | Authors: | Li, K.J, Li, X.J, Wu, B.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.
Front Microbiol, 13, 2022
|
|
5ZTY
 
 | | Crystal structure of human G protein coupled receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | Deposit date: | 2018-05-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
8GUS
 
 | | Cryo-EM structure of HU-CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUQ
 
 | | Cryo-EM structure of CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUT
 
 | | Cryo-EM structure of LEI-CB2-Gi complex | | Descriptor: | 1-[[4-[5-fluoranyl-6-[(oxan-4-ylamino)methyl]pyridin-2-yl]phenyl]methyl]-3-(2-methylpropyl)imidazolidine-2,4-dione, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, X.T, Chang, H, Wu, L.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUR
 
 | | Cryo-EM structure of CP-CB2-G protein complex | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8K2D
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
5VK0
 
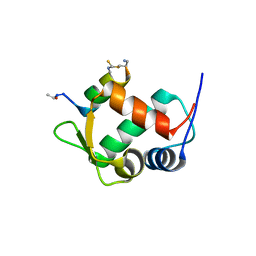 | |
5VK1
 
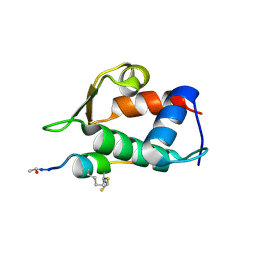 | |
4QRE
 
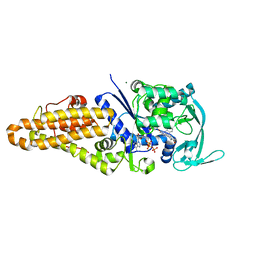 | |
7VU7
 
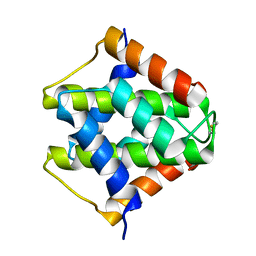 | |
8YMZ
 
 | | Structure of ZBTB43 in complex with CACA containing B-form DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*GP*TP*GP*TP*AP*TP*GP*TP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*AP*CP*AP*TP*AP*CP*AP*CP*AP*CP*T)-3'), ZINC ION, ... | | Authors: | Li, X, Yang, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the recognition of purine-pyrimidine dinucleotide repeats by zinc finger protein ZBTB43.
Febs J., 291, 2024
|
|
6IRL
 
 | |
6KX9
 
 | |
7CYA
 
 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYB
 
 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
8JQE
 
 | | Structure of CmCBDA in complex with Mn2+ and glycerol | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
8JQF
 
 | | Structure of CmCBDA in complex with Ni2+ and Glycerol | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
7VS7
 
 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
4TW2
 
 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
9M7V
 
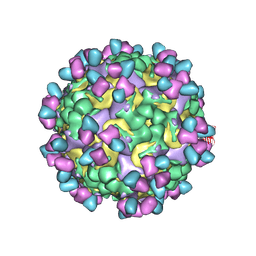 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab CT11F9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2025-03-11 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Development of In Vitro Potency Methods to Replace In Vivo Tests for Enterovirus 71 Inactivated Vaccine (Human Diploid Cell-Based/Vero Cell-Based).
Vaccines (Basel), 13, 2025
|
|
5XRR
 
 | | Crystal structure of FUS (54-59) SYSSYG | | Descriptor: | RNA-binding protein FUS, ZINC ION | | Authors: | Zhao, M, Gui, X, Li, D, Liu, C. | | Deposit date: | 2017-06-09 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7F3B
 
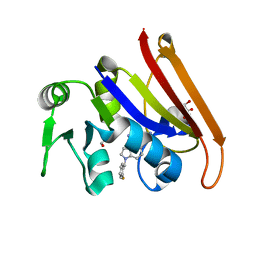 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
4CO5
 
 | |
