8Z3F
 
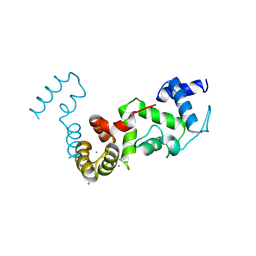 | | Complex structure of CIB2 and TMC1 CR1 | | Descriptor: | CALCIUM ION, Calcium and integrin-binding family member 2, Transmembrane channel-like protein 1 | | Authors: | Wu, S, Lin, L, Lu, Q. | | Deposit date: | 2024-04-15 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechano-electrical transduction components TMC1-CIB2 undergo a Ca 2+ -induced conformational change linked to hearing loss.
Dev.Cell, 60, 2025
|
|
7TTI
 
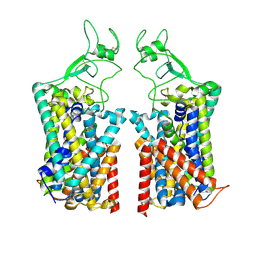 | | Human KCC1 bound with VU0463271 In an outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TTH
 
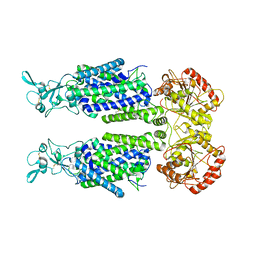 | | Human potassium-chloride cotransporter 1 in inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TQQ
 
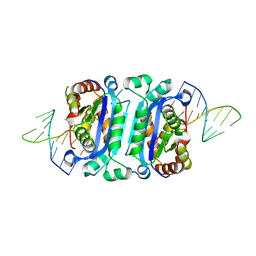 | | Structure of human TREX1-DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*GP*GP*CP*CP*GP*GP*CP*CP*AP*TP*C)-3'), Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
7TQO
 
 | |
7TQN
 
 | |
7TQP
 
 | | Structure of human TREX1 | | Descriptor: | SULFATE ION, Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
5NE4
 
 | |
5ISY
 
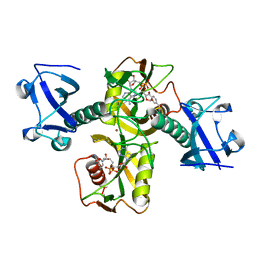 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
5NED
 
 | |
7CH5
 
 | |
5NEJ
 
 | |
5NET
 
 | |
5NEU
 
 | |
5NEM
 
 | |
7CHF
 
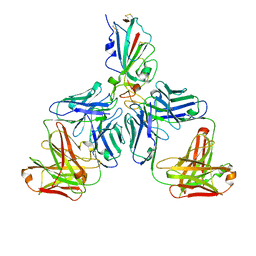 | |
5NER
 
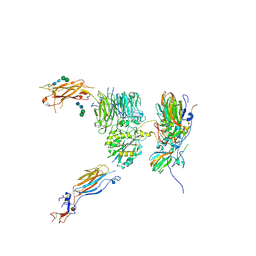 | |
7CHE
 
 | |
7CH4
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
6U9H
 
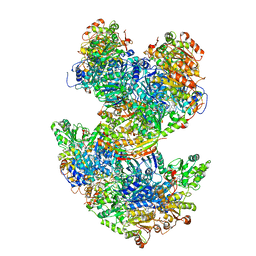 | | Arabidopsis thaliana acetohydroxyacid synthase complex | | Descriptor: | Acetolactate synthase small subunit 2, chloroplastic, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Low, Y.S, Rao, Z. | | Deposit date: | 2019-09-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of fungal and plant acetohydroxyacid synthases.
Nature, 586, 2020
|
|
6L8O
 
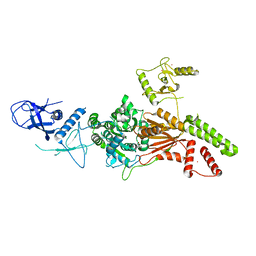 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
3HV9
 
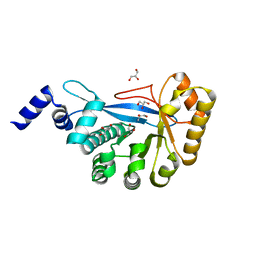 | | Crystal structure of FimX EAL domain from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Protein FimX | | Authors: | Navarro, M.V.A.S, De, N, Bae, N, Sondermann, H. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX.
Structure, 17, 2009
|
|
3HVA
 
 | |
