6MYO
 
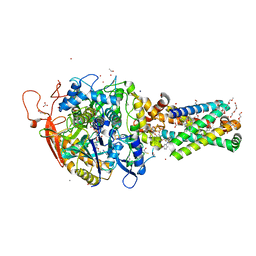 | | Avian mitochondrial complex II with flutolanyl bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
7BAJ
 
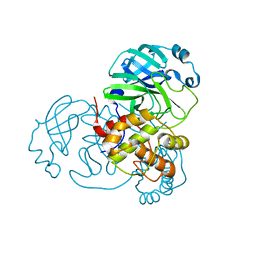 | |
7BAK
 
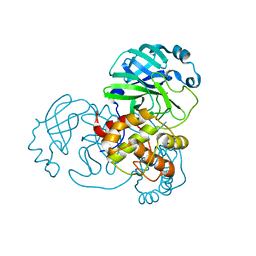 | |
7BAL
 
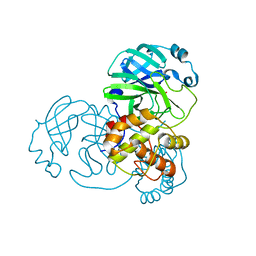 | |
5ZUD
 
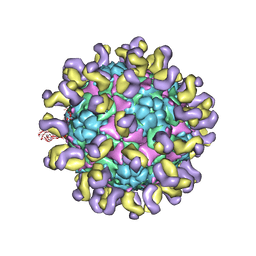 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZV7
 
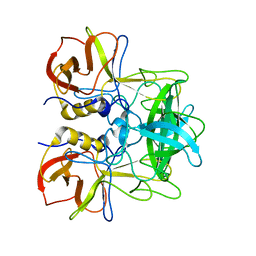 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZVC
 
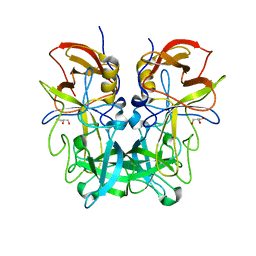 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV9
 
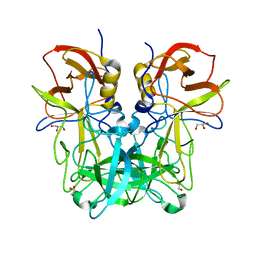 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUF
 
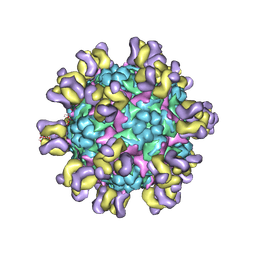 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZV5
 
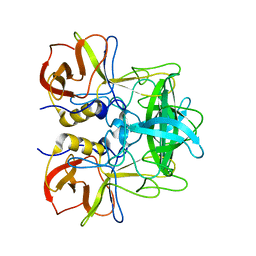 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZKQ
 
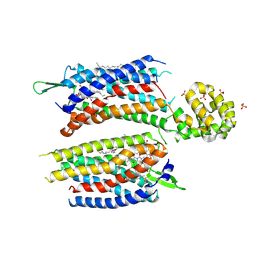 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZKP
 
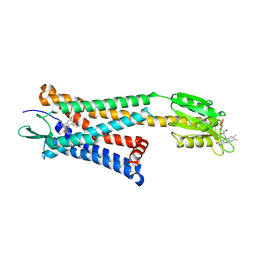 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZUQ
 
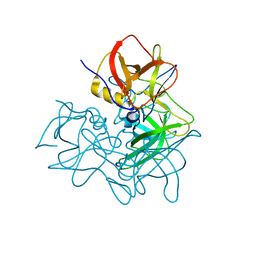 | | P domain of GII.17-1978 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUS
 
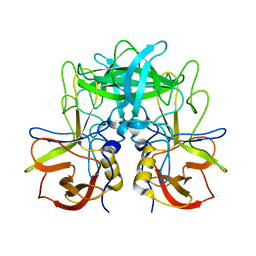 | | P domain of GII.17-2014/15 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
8HBK
 
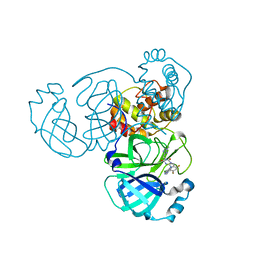 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M. | | Deposit date: | 2022-10-29 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
7DTE
 
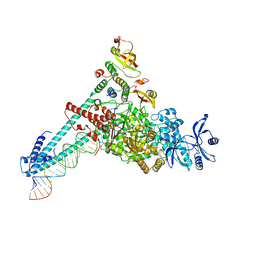 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
7BQA
 
 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
7EJU
 
 | | Junin virus(JUNV) RNA polymerase L complexed with Z protein | | Descriptor: | MAGNESIUM ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Chen, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2021-07-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for recognition and regulation of arenavirus polymerase L by Z protein.
Nat Commun, 12, 2021
|
|
7E9T
 
 | | Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhu, Y, Tai, L, Zhu, G, Yin, G, Sun, F. | | Deposit date: | 2021-03-05 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CAG
 
 | | Mycobacterium smegmatis LpqY-SugABC complex in the catalytic intermediate state | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
