3P65
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3P68
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3P64
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3P4Z
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-07 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
5YN8
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN6
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNN
 
 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNI
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNQ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNP
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNB
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
4JZR
 
 | | Structure of Prolyl Hydroxylase Domain-containing Protein (PHD) with Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(biphenyl-4-yl)-8-[(1-methyl-1H-imidazol-2-yl)methyl]-2,8-diazaspiro[4.5]decan-1-one, Egl nine homolog 1, ... | | Authors: | Ma, Y, Yang, L. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
9EV3
 
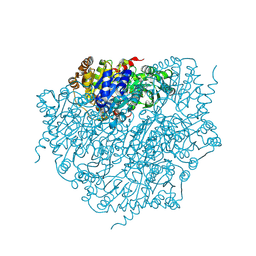 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO) purified from bacteria grown in acetate minimal medium | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
9EV5
 
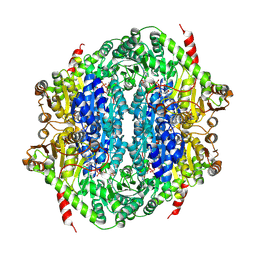 | | Corynebacterium glutamicum CS176 pyruvate:quinone oxidoreductase (PQO) in complex with FAD and thiamine diphosphate-magnesium ion | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
9EV4
 
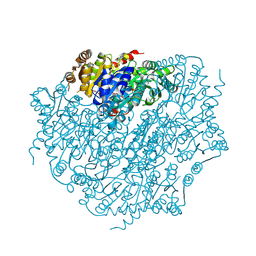 | | Pyruvate:quinone oxidoreductase (PQO) from Corynebacterium glutamicum CS176 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thiamine pyrophosphate-requiring enzymes [acetolactate synthase, pyruvate dehydrogenase (Cytochrome), ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
9EV6
 
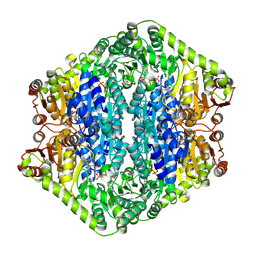 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO), C-terminal truncated construct | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
5YNO
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
8XK7
 
 | | binary complex of DNA polymerase SFM4-3 recognizing C2 methyoxy nucleotide | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA polymerase I, thermostable, ... | | Authors: | Wen, C, Liu, H, Yang, L, Gong, W. | | Deposit date: | 2023-12-22 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for C2'-methoxy Recognition by DNA Polymerases and Function Improvement.
J.Mol.Biol., 436, 2024
|
|
8XJR
 
 | | Apo form of DNA polymerase SFM4-3 recognizing C2 methyoxy nucleotide | | Descriptor: | DNA polymerase I, thermostable, SULFATE ION | | Authors: | Wen, C, Liu, H, Yang, L, Gong, W. | | Deposit date: | 2023-12-22 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for C2'-methoxy Recognition by DNA Polymerases and Function Improvement.
J.Mol.Biol., 436, 2024
|
|
