6E3S
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3T
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3U
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
7ZDV
 
 | |
7ZDA
 
 | |
7ZDR
 
 | |
7ZDF
 
 | |
7ZDT
 
 | |
7ZDB
 
 | |
7ZDE
 
 | |
7ZEC
 
 | |
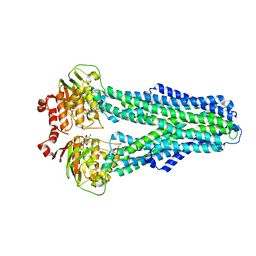
7ZDG
 
 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDS
 
 | |
7ZDW
 
 | |
7ZDL
 
 | |
7ZE5
 
 | |
7ZDK
 
 | |
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5WTI
 
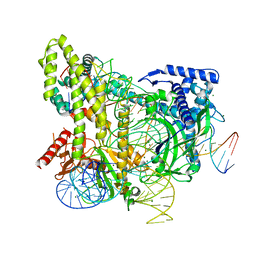 | | Crystal structure of the CRISPR-associated protein in complex with crRNA and DNA | | Descriptor: | CRISPR-associated protein, DNA (28-MER), DNA (5'-D(P*GP*TP*GP*TP*GP*GP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Wu, D, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2016-12-13 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA
Cell Res., 27, 2017
|
|
5XFF
 
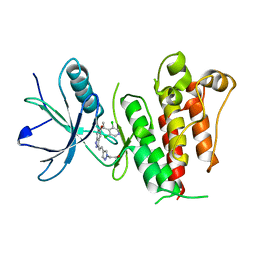 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550L) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
4M4X
 
 | |
5ZZ2
 
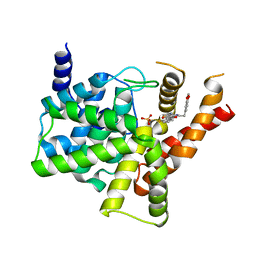 | | Crystal structure of PDE5 in complex with inhibitor LW1634 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
4DNW
 
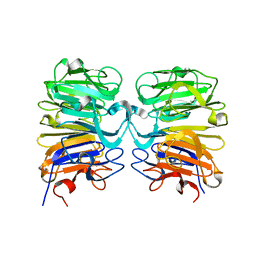 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
