5HZK
 
 | | Crystal structure of photoinhibitable Intersectin1 containing wildtype LOV2 domain in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
1O76
 
 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
5HZH
 
 | | Crystal structure of photoinhibitable Rac1 containing C450A mutant LOV2 domain | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
3MGX
 
 | |
1UYU
 
 | | Xenon COMPLEX OF wildtype P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, POTASSIUM ION, ... | | Authors: | Wade, R.C, Winn, P.J, Schlichting, I, Sudarko, X. | | Deposit date: | 2004-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Survey of Active Site Access Channels in Cytochromes P450
J.Inorg.Biochem., 98, 2004
|
|
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
3TEN
 
 | | Holo form of carbon disulfide hydrolase | | Descriptor: | CS2 hydrolase, ZINC ION | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
3TMK
 
 | | CRYSTAL STRUCTURE OF YEAST THYMIDYLATE KINASE COMPLEXED WITH THE BISUBSTRATE INHIBITOR TP5A AT 2.0 A RESOLUTION: IMPLICATIONS FOR CATALYSIS AND AZT ACTIVATION | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDYLATE KINASE | | Authors: | Lavie, A, Schlichting, I, Konrad, M, Goody, R.S, Brundiers, R, Reinstein, J. | | Deposit date: | 1998-01-26 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5'-adenosyl) P5-(5'-thymidyl) pentaphosphate (TP5A) at 2.0 A resolution: implications for catalysis and AZT activation.
Biochemistry, 37, 1998
|
|
8BKN
 
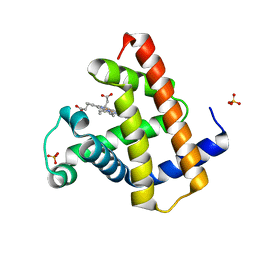 | |
8BKH
 
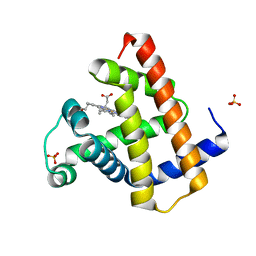 | |
3EJE
 
 | |
3EJD
 
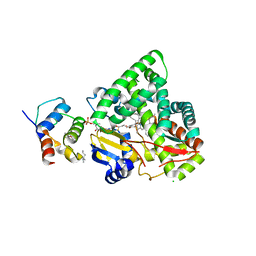 | |
3EJB
 
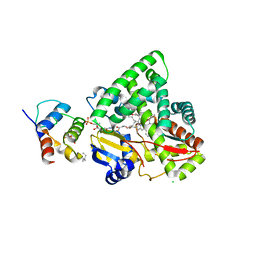 | |
1LTX
 
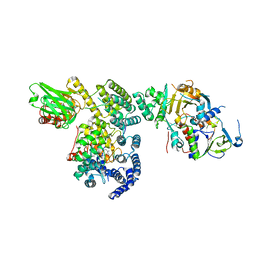 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8C
 
 | | Composite structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8A
 
 | | Crystal Structure of rsEGFP2 in the non-fluorescent off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
2J9X
 
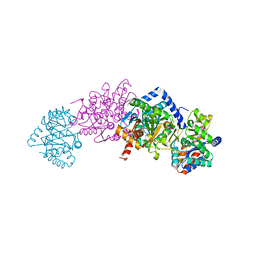 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
5OER
 
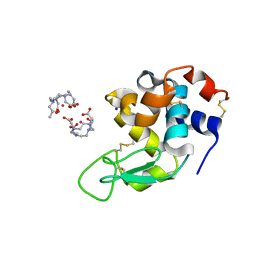 | | Hen egg-white lysozyme refined against 5000 9 keV diffraction patterns | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C, ... | | Authors: | Gorel, A, Schlichting, I. | | Deposit date: | 2017-07-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multi-wavelength anomalous diffraction de novo phasing using a two-colour X-ray free-electron laser with wide tunability.
Nat Commun, 8, 2017
|
|
3NC7
 
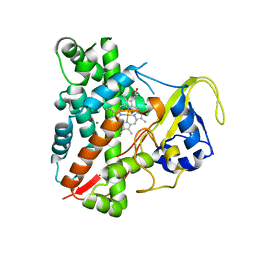 | | CYP134A1 2-phenylimidazole bound structure | | Descriptor: | 2-phenyl-1H-imidazole, Cytochrome P450 cypX, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
3NC5
 
 | | CYP134A1 structure with an open substrate binding loop | | Descriptor: | Cytochrome P450 cypX, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
3NC6
 
 | | CYP134A1 1-phenylimidazole bound structure | | Descriptor: | 1-phenyl-1H-imidazole, Cytochrome P450 cypX, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
3NC3
 
 | | CYP134A1 structure with a closed substrate binding loop | | Descriptor: | Cytochrome P450 cypX, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
