2C4P
 
 | |
2CLW
 
 | |
2CEO
 
 | | thyroxine-binding globulin complex with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, GLYCEROL, THYROXINE-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J, Carrell, R.W. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Mechanism for the Carriage and Release of Thyroxine in the Blood.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6HUF
 
 | | Coping with strong translational non-crystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Perez-Dorado, I, Murray, J.W, Tate, E.W, Cota, E, Read, R.J. | | Deposit date: | 2018-10-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Coping with strong translational noncrystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6I7K
 
 | | Crystal structure of monomeric FICD mutant L258D complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ETHANOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7J
 
 | | Crystal structure of monomeric FICD mutant L258D | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I3I
 
 | |
6I7H
 
 | | Crystal structure of dimeric FICD mutant K256S | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7I
 
 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I3F
 
 | | Crystal structure of the complex of human angiotensinogen and renin at 2.55 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, ... | | Authors: | Yan, Y, Read, R.J. | | Deposit date: | 2018-11-06 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specificity of renin-mediated angiotensinogen cleavage.
J. Biol. Chem., 294, 2019
|
|
6I7G
 
 | | Crystal structure of dimeric wild type FICD complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7L
 
 | | Crystal structure of monomeric FICD mutant L258D complexed with MgAMP-PNP | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
7PK0
 
 | |
7PK1
 
 | |
7PK2
 
 | |
1PRT
 
 | | THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN | | Descriptor: | PERTUSSIS TOXIN (SUBUNIT S1), PERTUSSIS TOXIN (SUBUNIT S2), PERTUSSIS TOXIN (SUBUNIT S3), ... | | Authors: | Stein, P.E, Read, R.J. | | Deposit date: | 1993-11-22 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of pertussis toxin.
Structure, 2, 1994
|
|
1QMB
 
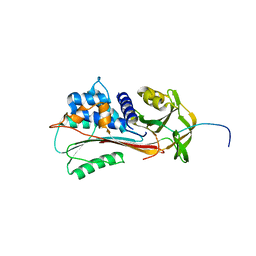 | | Cleaved alpha-1-antitrypsin polymer | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Huntington, J.A, Pannu, N.S, Hazes, B, Read, R.J, Lomas, D.A, Carrell, R.W. | | Deposit date: | 1999-09-24 | | Release date: | 2000-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A 2.6A Structure of a Serpin Polymer and Implications for Conformational Disease
J.Mol.Biol., 293, 1999
|
|
8AXG
 
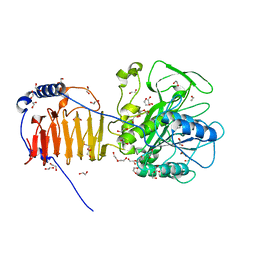 | | Crystal structure of Fusobacterium nucleatum fusolisin protease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fusolisin, ... | | Authors: | Isupov, M.N, Wiener, R, Rouvinski, A, Fahoum, J, Kumar, M, Read, R.J. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of Fusobacterium nucleatum fusolisin protease
To Be Published
|
|
6RPX
 
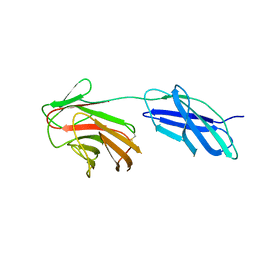 | |
6RPZ
 
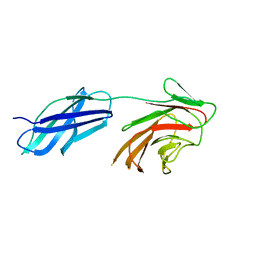 | |
6RPY
 
 | | Cytokine receptor-like factor 3 C-terminus residues 174-442: Hg-SAD derivative | | Descriptor: | Cytokine receptor-like factor 3, MERCURY (II) ION | | Authors: | Mifsud, R.W, Yan, Y, Bennett, C, Read, R.J. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-25 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CRLF3 plays a key role in the final stage of platelet genesis and is a potential therapeutic target for thrombocythemia.
Blood, 139, 2022
|
|
6EOE
 
 | | Crystal structure of AMPylated GRP78 with nucleotide | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EOB
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 1) | | Descriptor: | 78 kDa glucose-regulated protein, PHOSPHATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EOC
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EOF
 
 | | Crystal structure of AMPylated GRP78 in ADP state | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, Y, Preissler, S, Read, R.J, Ron, D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
