8S96
 
 | | RNase A-Adenosine 5'-Heptaphosphate (RNaseA.p7A) | | Descriptor: | Ribonuclease pancreatic, adenosine 5'-heptaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A
Acs Cent.Sci., 2024
|
|
2Q4G
 
 | | Ensemble refinement of the protein crystal structure of human ribonuclease inhibitor complexed with ribonuclease I | | Descriptor: | CITRIC ACID, Ribonuclease inhibitor, Ribonuclease pancreatic | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein.
J.Mol.Biol., 368, 2007
|
|
8GC9
 
 | | RNase A-Uridine 5'-Heptaphosphate (RNase A.p7U) | | Descriptor: | Ribonuclease pancreatic, uridine 5'-heptaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A
Acs Cent.Sci., 2024
|
|
8GGG
 
 | | RNase A-Adenosine 5'-Hexaphosphate (RNaseA.p6A) | | Descriptor: | GLYCEROL, Ribonuclease pancreatic, adenosine 5'-hexaphosphate | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A
Acs Cent.Sci., 2024
|
|
8FHM
 
 | | RNase A-Uridine 5'-Hexaphosphate (RNaseA.p6U) | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]uridine, Ribonuclease pancreatic | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-24 | | Method: | SOLUTION SCATTERING (1.79 Å), X-RAY DIFFRACTION | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A
Acs Cent.Sci., 2024
|
|
3FHK
 
 | | Crystal structure of APC1446, B.subtilis YphP disulfide isomerase | | Descriptor: | SULFATE ION, UPF0403 protein yphP | | Authors: | Derewenda, U, Boczek, T, Cooper, D.R, Yu, M, Hung, L, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-09 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif .
Biochemistry, 48, 2009
|
|
6VXE
 
 | |
6VXC
 
 | |
5AGX
 
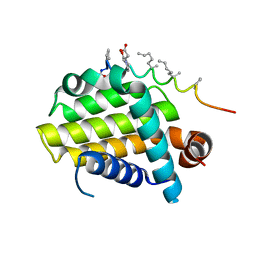 | | Bcl-2 alpha beta-1 LINEAR complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
5AGW
 
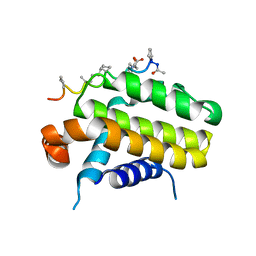 | | Bcl-2 alpha beta-1 complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
1N3Z
 
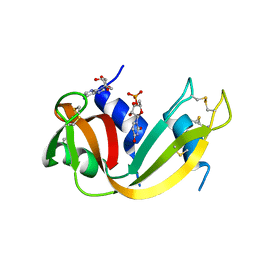 | | Crystal structure of the [S-carboxyamidomethyl-Cys31, S-carboxyamidomethyl-Cys32] monomeric derivative of the bovine seminal ribonuclease in the liganded state | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, ADENOSINE, Ribonuclease, ... | | Authors: | Sica, F, Di Fiore, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-10-30 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative
Proteins, 52, 2003
|
|
