2K6Q
 
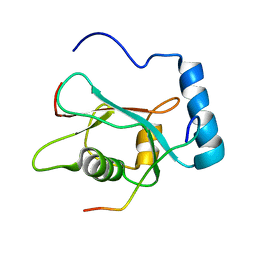 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1WSV
 
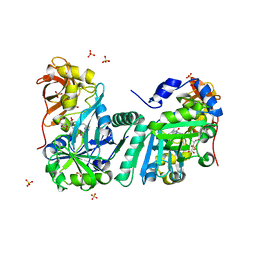 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1I6I
 
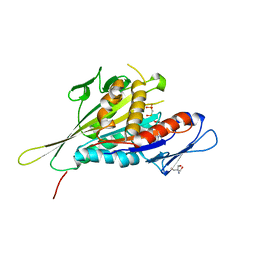 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-AMPPCP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1I5S
 
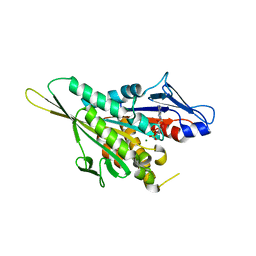 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-02-28 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1WSR
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1X2H
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1IR2
 
 | | Crystal Structure of Activated Ribulose-1,5-bisphosphate Carboxylase/oxygenase (Rubisco) from Green alga, Chlamydomonas reinhardtii Complexed with 2-Carboxyarabinitol-1,5-bisphosphate (2-CABP) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, Large subunit of Rubisco, ... | | Authors: | Mizohata, E, Matsumura, H, Okano, Y, Kumei, M, Takuma, H, Onodera, J, Kato, K, Shibata, N, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2001-09-03 | | Release date: | 2002-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.
J.Mol.Biol., 316, 2002
|
|
5YGY
 
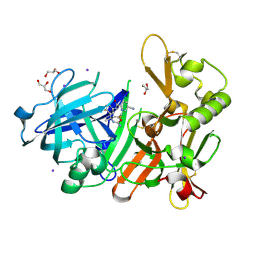 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
1IR1
 
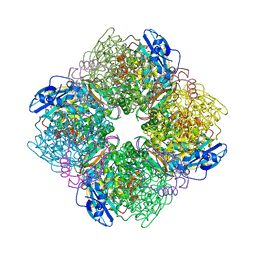 | | Crystal Structure of Spinach Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco) Complexed with CO2, Mg2+ and 2-Carboxyarabinitol-1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Large subunit of Rubisco, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Okano, Y, Kumei, M, Takuma, H, Onodera, J, Kato, K, Shibata, N, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2001-08-31 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.
J.Mol.Biol., 316, 2002
|
|
1J2F
 
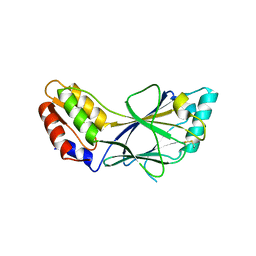 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
5XJE
 
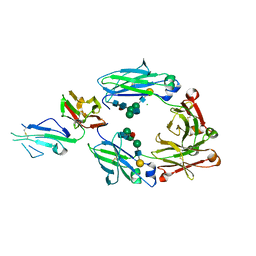 | | Crystal structure of fucosylated IgG1 Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5XJF
 
 | | Crystal structure of fucosylated IgG Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
7VIW
 
 | | Dark adapted MmCPDII during oxidized to semiquinone TR-SFX studies | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ3
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (400 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJC
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (100 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ2
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (125 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VIZ
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (250 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ9
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (semiquinone dark structure) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ1
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (10 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJK
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (100 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJB
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
