2GQL
 
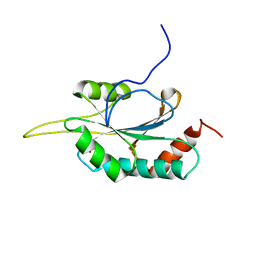 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GQK
 
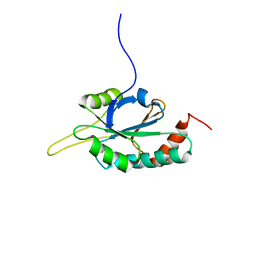 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HRF
 
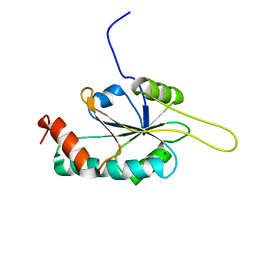 | | Solution Structure of Cu(I) P174L HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HRN
 
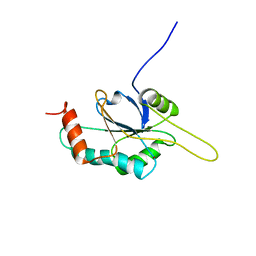 | | Solution Structure of Cu(I) P174L-HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GGT
 
 | | Crystal structure of human SCO1 complexed with nickel. | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | Deposit date: | 2006-03-24 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GQM
 
 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GVP
 
 | | Solution structure of Human apo Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Paulmaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT5
 
 | | Solution structure of apo Human Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT6
 
 | | Solution structure of Human Cu(I) Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3QKL
 
 | | Spirochromane Akt Inhibitors | | Descriptor: | GSK-3 beta peptide, N-{(2S)-3-[(3S)-8',9'-dihydro-1H,3'H-spiro[piperidine-3,7'-pyrano[3,2-e]indazol]-1-yl]-2-hydroxypropyl}-N-(2-ethoxyethyl)-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QKK
 
 | | Spirochromane Akt Inhibitors | | Descriptor: | GSK-3 beta peptide, N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(2R)-6-hydroxy-4-oxo-3,4-dihydro-1'H-spiro[chromene-2,3'-piperidin]-1'-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QKM
 
 | | Spirocyclic sulfonamides as AKT inhibitors | | Descriptor: | N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(5R)-2-(quinazolin-4-yl)-2,7-diazaspiro[4.5]dec-7-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Xu, R, Banka, A, Blake, J.F, Mitchell, I.S, Wallace, E.M, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of spirocyclic sulfonamides as potent Akt inhibitors with exquisite selectivity against PKA.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1FG4
 
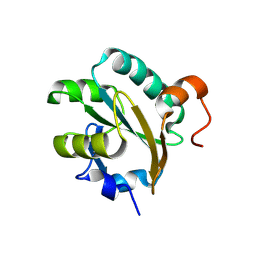 | | STRUCTURE OF TRYPAREDOXIN II | | Descriptor: | TRYPAREDOXIN II | | Authors: | Hofmann, B, Budde, H, Bruns, K, Guerrero, S.A, Kalisz, H.M, Menge, U, Montemartini, M, Nogoceke, E, Steinert, P, Wissing, J.B, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-07-28 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1EWX
 
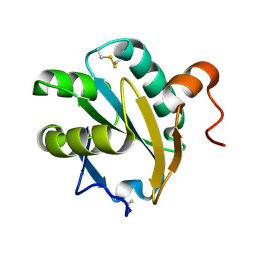 | | Crystal structure of native tryparedoxin I from Crithidia fasciculata | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-04-28 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1EZK
 
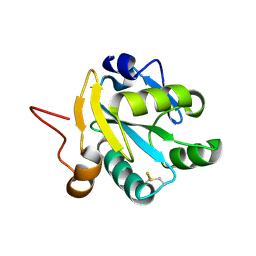 | | Crystal structure of recombinant tryparedoxin I | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
2KDU
 
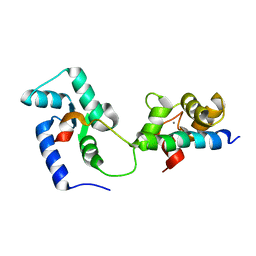 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
2N6J
 
 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
5OHE
 
 | | Globin sensor domain of AfGcHK (FeIII form) in complex with cyanide | | Descriptor: | CHLORIDE ION, CYANIDE ION, Globin-coupled histidine kinase, ... | | Authors: | Skalova, T, Kolenko, P, Dohnalek, J, Stranava, M, Martinkova, M. | | Deposit date: | 2017-07-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction.
J. Biol. Chem., 292, 2017
|
|
5OHF
 
 | | Globin sensor domain of AfGcHK (FeIII form) in complex with cyanide, partially reduced | | Descriptor: | CHLORIDE ION, CYANIDE ION, Globin-coupled histidine kinase, ... | | Authors: | Skalova, T, Kolenko, P, Dohnalek, J, Stranava, M, Martinkova, M. | | Deposit date: | 2017-07-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction.
J. Biol. Chem., 292, 2017
|
|
4DMW
 
 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | Descriptor: | Toxin A | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4BZO
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone inhibitor | | Descriptor: | N-[(1S)-2-AMINO-1-PHENYLETHYL]-2-[(4S)-7-(2-FLUORO-4-PYRIDINYL)-1-OXO-1,2,3,4-TETRAHYDROPYRROLO[1,2-A]PYRAZIN-4-YL]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
4BZN
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo(1,2-a)Pyrazinone inhibitor | | Descriptor: | N-(2,2-dimethylpropyl)-2-[1-oxo-7-(thiophen-3-yl)-1,2,3,4-tetrahydropyrrolo[1,2-a]pyrazin-4-yl]acetamide, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
4Q30
 
 | | Nitrowillardiine bound to the ligand binding domain of GluA2 at pH 3.5 | | Descriptor: | 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, Glutamate receptor 2 CHIMERIC PROTEIN, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thermodynamics and mechanism of the interaction of willardiine partial agonists with a glutamate receptor: implications for drug development.
Biochemistry, 53, 2014
|
|
4NAU
 
 | | S. aureus CoaD with Inhibitor | | Descriptor: | 2-[2-[(1S,2S)-2-[(3,4-dichlorophenyl)methylcarbamoyl]cyclohexyl]-6-ethyl-pyrimidin-4-yl]-4-oxidanyl-6-oxidanylidene-1H-pyrimidine-5-carboxamide, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phosphopantetheine adenylyltransferase | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Inhibitors of 4'-Phosphopantetheine Adenylyltransferase (PPAT) To Validate PPAT as a Target for Antibacterial Therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
