1F9J
 
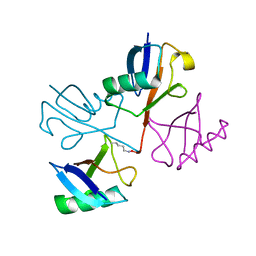 | | STRUCTURE OF A NEW CRYSTAL FORM OF TETRAUBIQUITIN | | Descriptor: | TETRAUBIQUITIN | | Authors: | Phillips, C.L, Thrower, J, Pickart, C.M, Hill, C.P. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a new crystal form of tetraubiquitin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3BIP
 
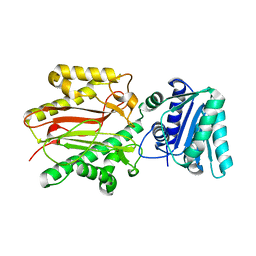 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIT
 
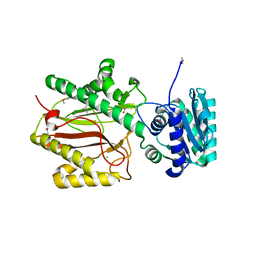 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FACT complex subunit SPT16, ... | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIQ
 
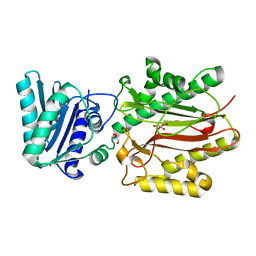 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16, GLYCEROL | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BZK
 
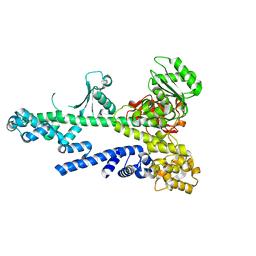 | |
3C3Q
 
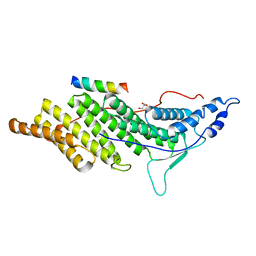 | | ALIX Bro1-domain:CHMIP4B co-crystal structure | | Descriptor: | Charged multivesicular body protein 4b peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BZC
 
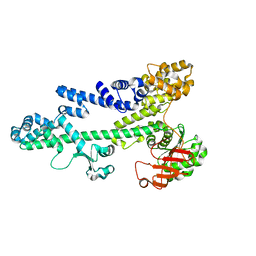 | |
3C3R
 
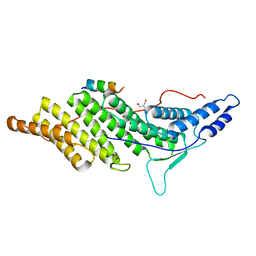 | | ALIX BRO1 CHMP4C complex | | Descriptor: | Charged multivesicular body protein 4c peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C3O
 
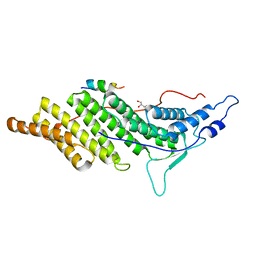 | | ALIX Bro1-domain:CHMIP4A co-crystal structure | | Descriptor: | Charged multivesicular body protein 4a peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1LC3
 
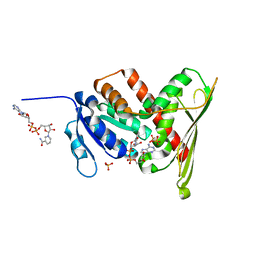 | | Crystal Structure of a Biliverdin Reductase Enzyme-Cofactor Complex | | Descriptor: | Biliverdin Reductase A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Whitby, F.G, Phillips, J.D, Hill, C.P, McCoubrey, W, Maines, M.D. | | Deposit date: | 2002-04-05 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex.
J.Mol.Biol., 319, 2002
|
|
1LC0
 
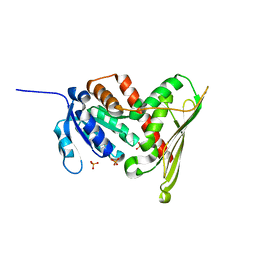 | | Structure of Biliverdin Reductase and the Enzyme-NADH Complex | | Descriptor: | Biliverdin Reductase A, PHOSPHATE ION | | Authors: | Whitby, F.G, Phillips, J.D, Hill, C.P, McCoubrey, W, Maines, M.D. | | Deposit date: | 2002-04-04 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex.
J.Mol.Biol., 319, 2002
|
|
1JR2
 
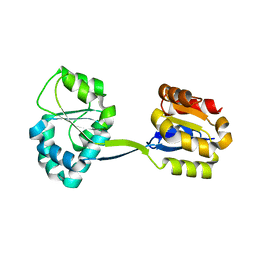 | | Structure of Uroporphyrinogen III Synthase | | Descriptor: | UROPORPHYRINOGEN-III SYNTHASE | | Authors: | Mathews, M.A, Schubert, H.L, Whitby, F.G, Alexander, K.J, Schadick, K, Bergonia, H.A, Phillips, J.D, Hill, C.P. | | Deposit date: | 2001-08-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of human uroporphyrinogen III synthase.
EMBO J., 20, 2001
|
|
1UCH
 
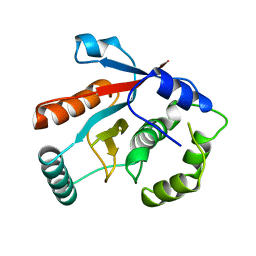 | | DEUBIQUITINATING ENZYME UCH-L3 (HUMAN) AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | UBIQUITIN C-TERMINAL HYDROLASE UCH-L3 | | Authors: | Johnston, S.C, Larsen, C.N, Cook, W.J, Wilkinson, K.D, Hill, C.P. | | Deposit date: | 1997-10-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution.
EMBO J., 16, 1997
|
|
1S1Q
 
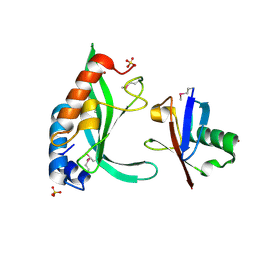 | | TSG101(UEV) domain in complex with Ubiquitin | | Descriptor: | ACETIC ACID, COPPER (II) ION, SULFATE ION, ... | | Authors: | Sundquist, W.I, Schubert, H.L, Kelly, B.N, Hill, G.C, Holton, J.M, Hill, C.P. | | Deposit date: | 2004-01-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin recognition by the human TSG101 protein
Mol.Cell, 13, 2004
|
|
5WHG
 
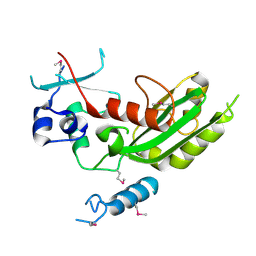 | | Vms1 mitochondrial localization core | | Descriptor: | Protein VMS1, ZINC ION | | Authors: | Fredrickson, E.K, Schubert, H.L, Rutter, J, Hill, C.P. | | Deposit date: | 2017-07-17 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sterol Oxidation Mediates Stress-Responsive Vms1 Translocation to Mitochondria.
Mol. Cell, 68, 2017
|
|
1XWI
 
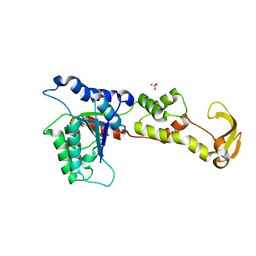 | |
5VKL
 
 | | SPT6 tSH2-RPB1 1476-1500 pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
3EIE
 
 | | Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state | | Descriptor: | SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
3JSE
 
 | |
3FRR
 
 | |
3L37
 
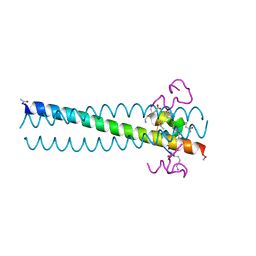 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
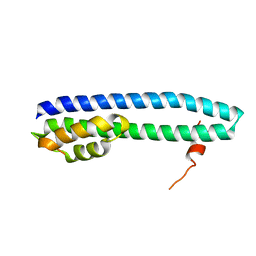
3FRT
 
 | |
3JRM
 
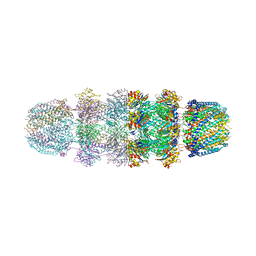 | |
3JTL
 
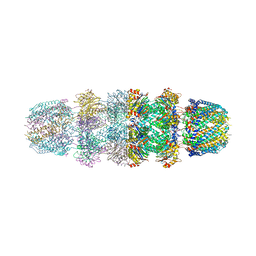 | |
3L36
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
