7UKD
 
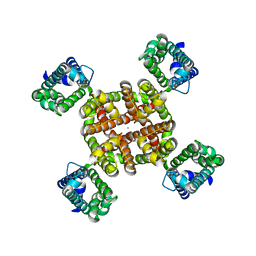 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
6VWS
 
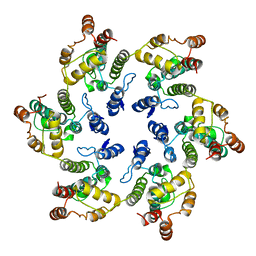 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
6W1S
 
 | | Atomic model of the mammalian Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Young, N, Asturias, F. | | Deposit date: | 2020-03-04 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
Cell, 178, 2019
|
|
5U93
 
 | |
5U9E
 
 | |
2P4H
 
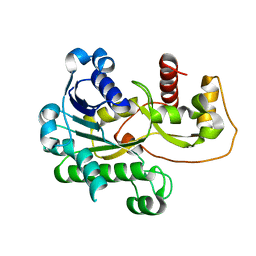 | |
3CNS
 
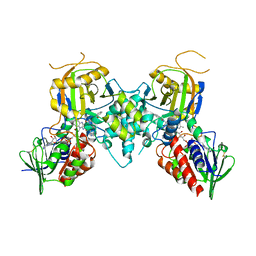 | |
1EEG
 
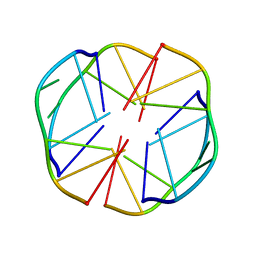 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
4KPY
 
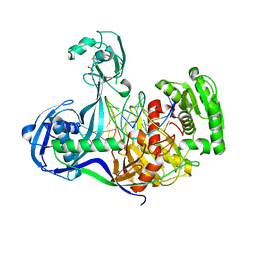 | | DNA binding protein and DNA complex structure | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6WKV
 
 | | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE) | | Descriptor: | Encapsulin, FLAVIN MONONUCLEOTIDE | | Authors: | Williams, E, Jenkins, M, Zhao, H, Juneja, P, Lutz, S. | | Deposit date: | 2020-04-17 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE)
To Be Published
|
|
6P05
 
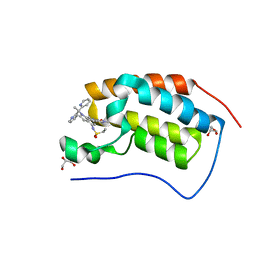 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with compound 27 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Zhao, J, Gutgesell, L.M, Shen, Z, Dye, K, Dubrovyskyii, O, Zhao, H, Huang, F, Tonetti, D.A, Thatcher, G.R. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
8Z9R
 
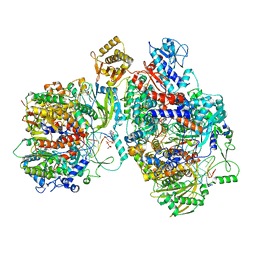 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8N
 
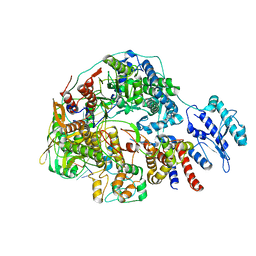 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z97
 
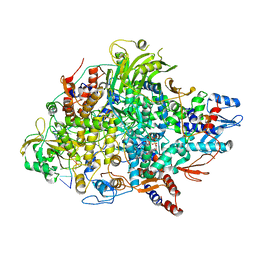 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8J
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8CEG
 
 | |
2FGT
 
 | | Crystal Structure of YycH from Bacillus subtilis | | Descriptor: | Two-component system yycF/yycG regulatory protein yycH | | Authors: | Szurmant, H, Zhao, H, Mohan, M.A, James, H.A, Varughese, K.I. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of YycH involved in the regulation of the essential YycFG two-component system in Bacillus subtilis reveals a novel tertiary structure.
Protein Sci., 15, 2006
|
|
1MSI
 
 | |
8EX5
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
