8IX1
 
 | | Cryo-EM structure of protonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
7CYC
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
6XCJ
 
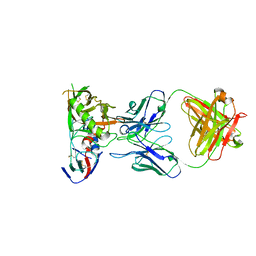 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
6XRT
 
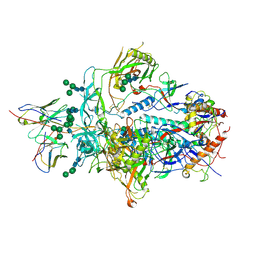 | |
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HVI
 
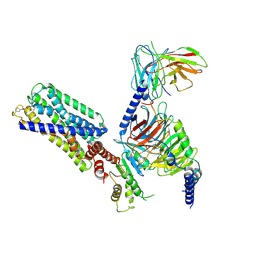 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
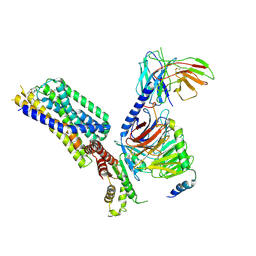 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8X8Q
 
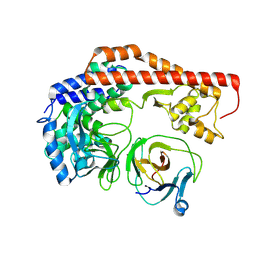 | | Structure of enterovirus protease in complex host factor | | Descriptor: | 2A protein (Fragment), Actin-histidine N-methyltransferase, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-28 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
8X77
 
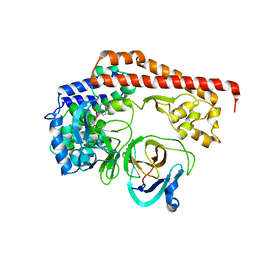 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
4FC7
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
8HQE
 
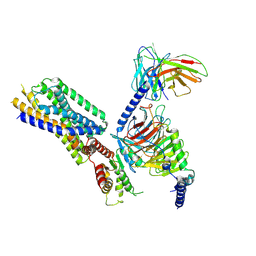 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
6LE0
 
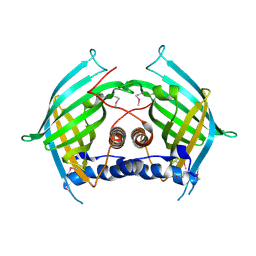 | |
2MK5
 
 | | Solution structure of a protein domain | | Descriptor: | Endolysin | | Authors: | Feng, Y, Gu, J. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
4OLS
 
 | | The amidase-2 domain of LysGH15 | | Descriptor: | Endolysin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
4OLK
 
 | | The CHAP domain of LysGH15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endolysin | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
4Y5U
 
 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VIL
 
 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 6) | | Descriptor: | 2-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-5-sulfamoylbenzamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VIO
 
 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 13) | | Descriptor: | 4-methoxy-N~1~-methyl-N~3~-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}benzene-1,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
8IL0
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 | | Descriptor: | Glycosyltransferase | | Authors: | Dai, Y, Li, P, Qiao, H, Xia, M, Liu, W, Fang, P. | | Deposit date: | 2023-03-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
5D39
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
