3FAU
 
 | | Crystal Structure of human small-MutS related domain | | Descriptor: | NEDD4-binding protein 2 | | Authors: | Kim, T.G, Kwon, T.H, Ryu, E.K, Min, K, Heo, S.-D, Song, K.M, Jun, W.J, Jung, E. | | Deposit date: | 2008-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Strcutral Dynamincs of the Endonuclease Small-MutS Related Domains of BCL3 binding protein
To be Published
|
|
2V59
 
 | |
2V58
 
 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5K59
 
 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
3EQB
 
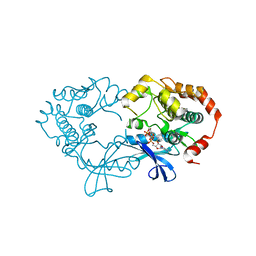 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2V0K
 
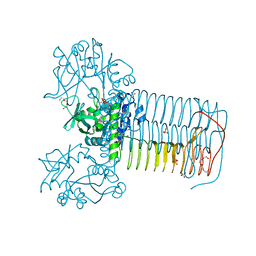 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0L
 
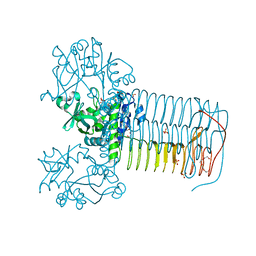 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0I
 
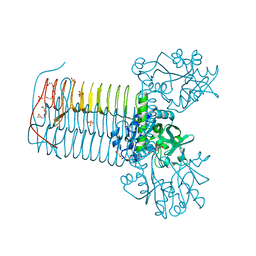 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0J
 
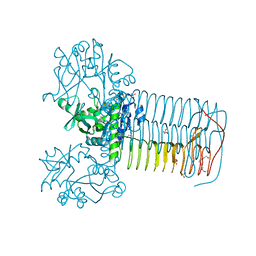 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, BIFUNCTIONAL PROTEIN GLMU, MAGNESIUM ION, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
2V0H
 
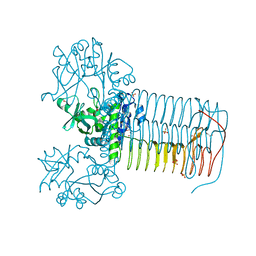 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
8JI1
 
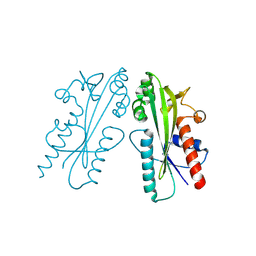 | | Crystal structure of Ham1 from Plasmodium falciparum | | Descriptor: | Inosine triphosphate pyrophosphatase | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of nucleotide housekeeping protein HAM1 from human malaria parasite Plasmodium falciparum.
Febs J., 291, 2024
|
|
7RRG
 
 | |
7L1B
 
 | |
7L1D
 
 | |
7L1C
 
 | |
4N1D
 
 | | Nodal/BMP2 chimera NB250 | | Descriptor: | Nodal/BMP2 chimera protein | | Authors: | Esquivies, L. | | Deposit date: | 2013-10-04 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Designer Nodal/BMP2 Chimeras Mimic Nodal Signaling, Promote Chondrogenesis, and Reveal a BMP2-like Structure.
J.Biol.Chem., 289, 2014
|
|
1AYK
 
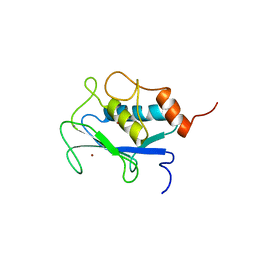 | |
2AYK
 
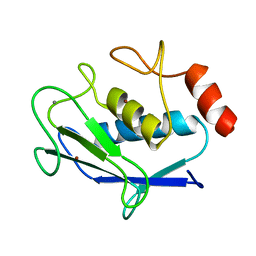 | |
8FFZ
 
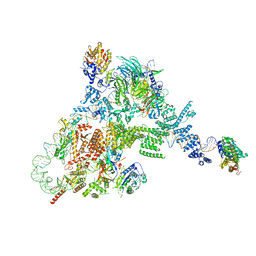 | | TFIIIA-TFIIIC-Brf1-TBP complex bound to 5S rRNA gene | | Descriptor: | DNA (151-MER), Transcription factor IIIA, Transcription factor IIIB 70 kDa subunit,TATA-box-binding protein, ... | | Authors: | Talyzina, A, He, Y. | | Deposit date: | 2022-12-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Mol.Cell, 83, 2023
|
|
2V5A
 
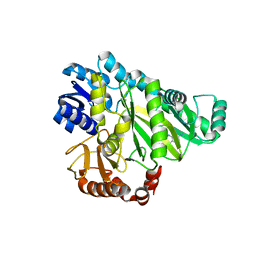 | |
3AYK
 
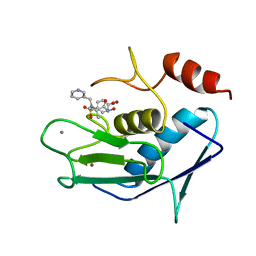 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
2N9H
 
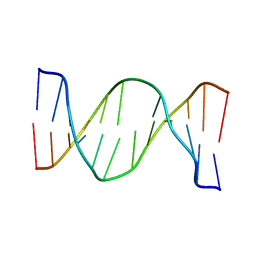 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9F
 
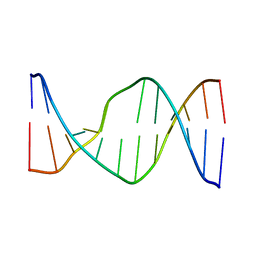 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3DY7
 
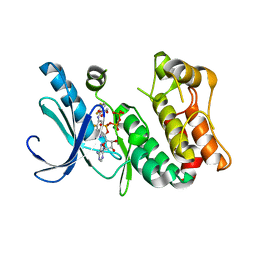 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | (5S)-4,5-difluoro-6-[(2-fluoro-4-iodophenyl)imino]-N-(2-hydroxyethoxy)cyclohexa-1,3-diene-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-07-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6EXZ
 
 | | Crystal structure of Mex67 C-term | | Descriptor: | FORMIC ACID, mRNA export factor MEX67 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mip6 binds directly to the Mex67 UBA domain to maintain low levels of Msn2/4 stress-dependent mRNAs.
Embo Rep., 2019
|
|
