6MYI
 
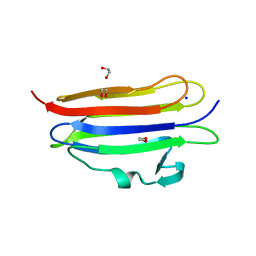 | | Pleurotus ostreatus OstreolysinA | | Descriptor: | 1,2-ETHANEDIOL, Ostreolysin A6, SODIUM ION | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
3BND
 
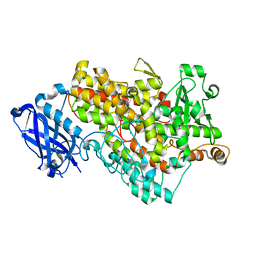 | | Lipoxygenase-1 (Soybean), I553V Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BNE
 
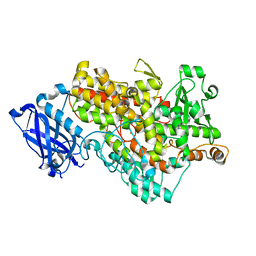 | | Lipoxygenase-1 (Soybean) I553A Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BNB
 
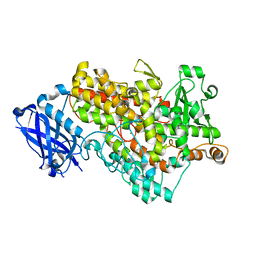 | | Lipoxygenase-1 (Soybean) I553L Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BNC
 
 | | Lipoxygenase-1 (Soybean) I553G Mutant | | Descriptor: | ACETIC ACID, FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6VVC
 
 | |
6VVE
 
 | | Legionella pneumophila Lpg2603 kinase bound to IP6, Mn2+, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dot/Icm T4SS effector, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Park, B.C. | | Deposit date: | 2020-02-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ALegionellaeffector kinase is activated by host inositol hexakisphosphate.
J.Biol.Chem., 295, 2020
|
|
6VVD
 
 | |
4PKY
 
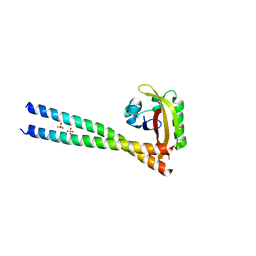 | | ARNT/HIF transcription factor/coactivator complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, SULFATE ION, ... | | Authors: | Tomchick, D.R, Partch, C.L, Gardner, K.H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coiled-coil Coactivators Play a Structural Role Mediating Interactions in Hypoxia-inducible Factor Heterodimerization.
J.Biol.Chem., 290, 2015
|
|
4R39
 
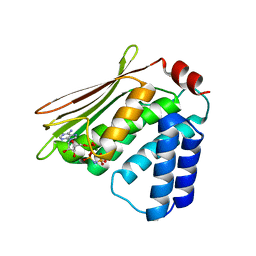 | |
4R3A
 
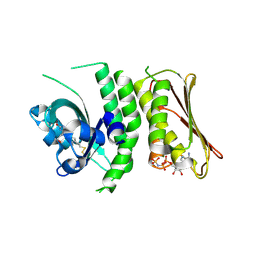 | | Erythrobacter litoralis EL346 blue-light activated histidine kinase | | Descriptor: | Blue-light-activated histidine kinase 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tomchick, D.R, Rivera-Cancel, G, Gardner, K.H. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R8Q
 
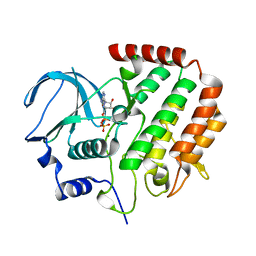 | |
4M63
 
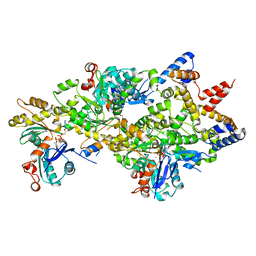 | | Crystal Structure of a Filament-Like Actin Trimer Bound to the Bacterial Effector VopL | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Zahm, J.A, Rosen, M.K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | The Bacterial Effector VopL Organizes Actin into Filament-like Structures.
Cell(Cambridge,Mass.), 155, 2013
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UF7
 
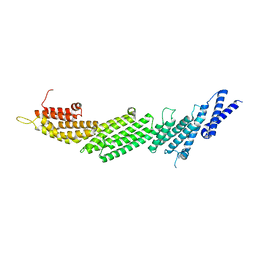 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
4IFW
 
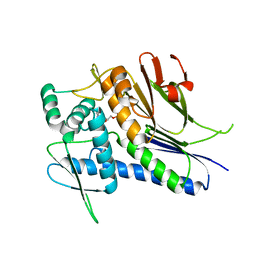 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, ADP inhibited form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4IG1
 
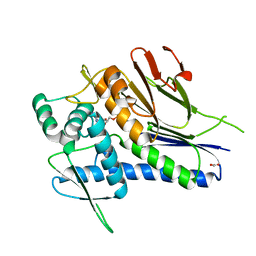 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mg(II)-AMP product bound form | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4318 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4IFU
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8334 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4IFX
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, FAD substrate bound form | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4IFZ
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mn(II)-AMP product bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9012 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
2QYF
 
 | |
4K6J
 
 | | Human cohesin inhibitor WapL | | Descriptor: | ACETATE ION, SULFATE ION, Wings apart-like protein homolog | | Authors: | Tomchick, D.R, Yu, H, Ouyang, Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6205 Å) | | Cite: | Structure of the human cohesin inhibitor Wapl.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
7M7A
 
 | | Legionella pneumophila MavQ lipid kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoinositide 3-kinase MavQ | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Hsieh, T.S, Lopez, V.A. | | Deposit date: | 2021-03-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dynamic remodeling of host membranes by self-organizing bacterial effectors.
Science, 372, 2021
|
|
7LFM
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
