2XFB
 
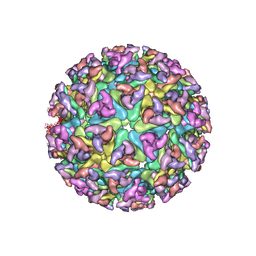 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SINDBIS VIRUS cryo- EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
3UYP
 
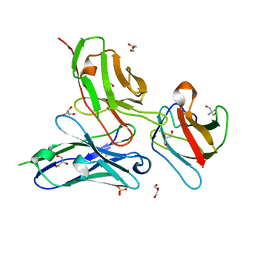 | | Crystal structure of the dengue virus serotype 4 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Envelope protein, GLYCEROL, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UZE
 
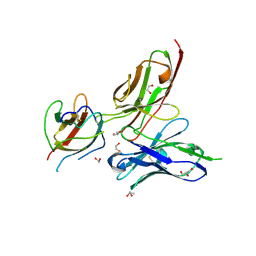 | | Crystal structure of the dengue virus serotype 3 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Envelope protein, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UZQ
 
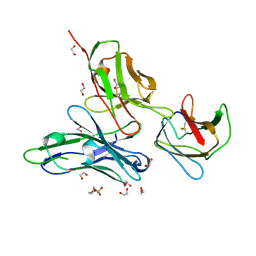 | | Crystal structure of the dengue virus serotype 1 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
3UZV
 
 | | Crystal structure of the dengue virus serotype 2 envelope protein domain III in complex with the variable domains of Mab 4E11 | | Descriptor: | ETHANOL, anti-dengue Mab 4E11, envelope protein | | Authors: | Cockburn, J.J.B, Navarro Sanchez, M.E, Fretes, N, Urvoas, A, Staropoli, I, Kikuti, C.M, Coffey, L.L, Arenzana Seisdedos, F, Bedouelle, H, Rey, F.A. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Structure, 20, 2012
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5FGC
 
 | |
6E18
 
 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
6ESC
 
 | | Crystal structure of Pseudorabies virus glycoprotein B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, GLYCEROL | | Authors: | Backovic, M, Vaney, M.C, Rey, F.A, Haouz, A. | | Deposit date: | 2017-10-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Function Dissection of Pseudorabies Virus Glycoprotein B Fusion Loops.
J.Virol., 92, 2018
|
|
6EGU
 
 | |
4UT6
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 B7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 B7, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5LBS
 
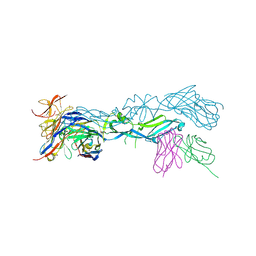 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
2BUO
 
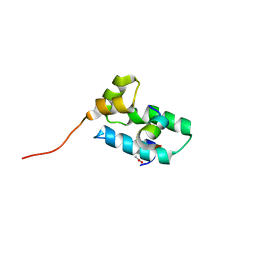 | | HIV-1 capsid C-terminal domain in complex with an inhibitor of particle assembly | | Descriptor: | ACETIC ACID, HIV-1 CAPSID PROTEIN, INHIBITOR OF CAPSID ASSEMBLY | | Authors: | Ternois, F, Sticht, J, Duquerroy, S, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The HIV-1 Capsid Protein C-Terminal Domain in Complex with a Virus Assembly Inhibitor
Nat.Struct.Mol.Biol., 12, 2005
|
|
2ALA
 
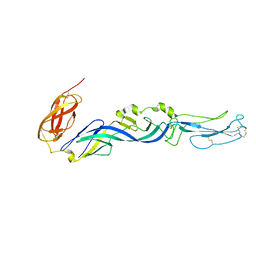 | | Crystal structure of the Semliki Forest Virus envelope protein E1 in its monomeric conformation. | | Descriptor: | Structural polyprotein (P130) | | Authors: | Roussel, A, Lescar, J, Vaney, M.C, Wengler, G, Wengler, G, Rey, F.A. | | Deposit date: | 2005-08-05 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus.
Structure, 14, 2006
|
|
7QRG
 
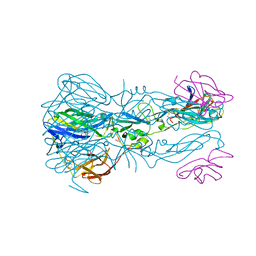 | |
1RER
 
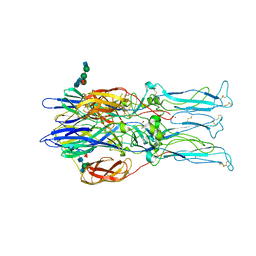 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
6AVQ
 
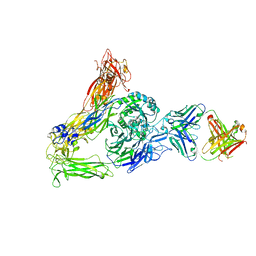 | | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha-V beta-3 Integrin via Steric Hindrance | | Descriptor: | Integrin alpha-V, Integrin beta-3, LM609 Fab heavy chain, ... | | Authors: | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
6AVR
 
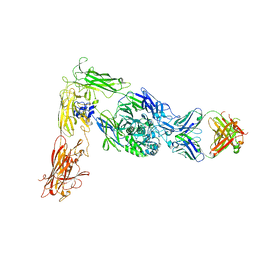 | | Human alpha-V beta-3 Integrin (intermediate conformation) in complex with the therapeutic antibody LM609 | | Descriptor: | Fab LM609 heavy chain, Fab LM609 light chain, Integrin alpha-V, ... | | Authors: | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
6AVU
 
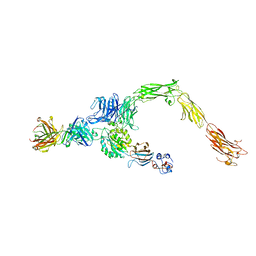 | | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | | Descriptor: | Fab LM609 heavy chain, Fab LM609 light chain, Integrin alpha-V, ... | | Authors: | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
6B3O
 
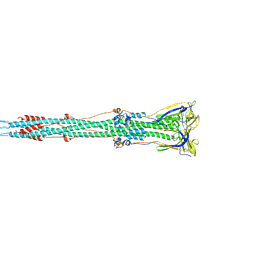 | | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Snijder, J, Xiong, X, Bosch, B.J, Rey, F.A, Veesler, D. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BFU
 
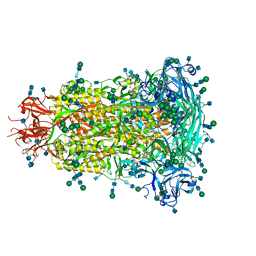 | | Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein, ... | | Authors: | Xiong, X, Tortorici, M.A, Snijder, S, Yoshioka, C, Walls, A.C, Li, W, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Rey, F.A, Bosch, B.J, Veesler, D. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Glycan Shield and Fusion Activation of a Deltacoronavirus Spike Glycoprotein Fine-Tuned for Enteric Infections.
J. Virol., 92, 2018
|
|
3IDE
 
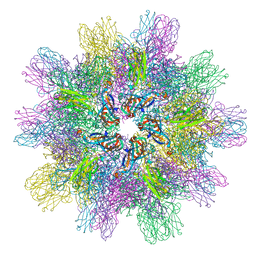 | | Structure of IPNV subviral particle | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Capsid protein VP2 | | Authors: | Coulibaly, F, Chevalier, C, Delmas, B, Rey, F.A. | | Deposit date: | 2009-07-21 | | Release date: | 2009-12-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of an aquabirnavirus particle: insights into antigenic diversity and virulence determinism
J.Virol., 84, 2010
|
|
3JCL
 
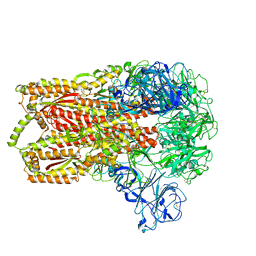 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
5N0A
 
 | | Crystal structure of A259C covalently linked dengue 2 virus envelope glycoprotein dimer in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 HEAVY CHAIN, Envelope Glycoprotein E, ... | | Authors: | Vaney, M.C, Rouvinski, A, Guardado-Calvo, P, Sharma, A, Rey, F.A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope.
Nat Commun, 8, 2017
|
|
6NB8
 
 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
