3MU8
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3K8J
 
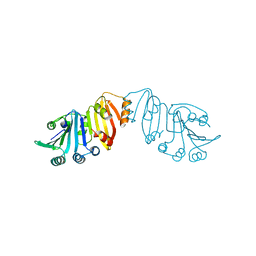 | | Structure of crystal form III of TP0453 | | Descriptor: | 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
3K5V
 
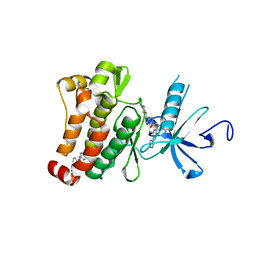 | | Structure of Abl kinase in complex with imatinib and GNF-2 | | Descriptor: | 3-(6-{[4-(trifluoromethoxy)phenyl]amino}pyrimidin-4-yl)benzamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, ... | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Rummel, G, Strauss, A. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors.
Nature, 463, 2010
|
|
3K8H
 
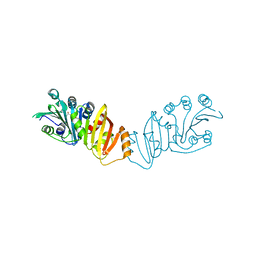 | | Structure of crystal form I of TP0453 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
5IWA
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with the GE81112 peptide antibiotic | | Descriptor: | (2S,3S)-2-{[(2S)-3-(2-amino-1H-imidazol-5-yl)-2-{[(2S,4S)-5-(carbamoyloxy)-4-hydroxy-2-({[(2S,3S)-3-hydroxypiperidin-2-yl]carbonyl}amino)pentanoyl]amino}propanoyl]amino}-3-(2-chloro-1H-imidazol-5-yl)-3-hydroxypropanoic acid, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Chieko, N, Masahito, K, Takemoto, C, Yokoyama, S, Connell, S.R, Fucini, P. | | Deposit date: | 2016-03-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6B0E
 
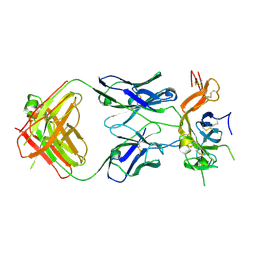 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1260 | | Descriptor: | 1260 antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, McLeod, B, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
1JJP
 
 | | A(GGGG) Pentad-Containing Dimeric DNA Quadruplex Involving Stacked G(anti)G(anti)G(anti)G(syn) Tetrads | | Descriptor: | 5'-D(*GP*GP*GP*AP*GP*GP*TP*TP*TP*GP*GP*GP*AP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads.
J.Mol.Biol., 311, 2001
|
|
5GZV
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5K59
 
 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
5KAX
 
 | | The structure of CTR107 protein bound to RHODAMINE 6G | | Descriptor: | CTR107 protein, GLYCEROL, RHODAMINE 6G | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8ANT
 
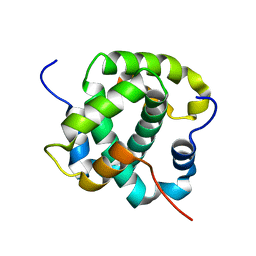 | |
8ANU
 
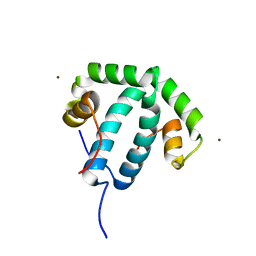 | | Crystal structure of protein phi3T-93 | | Descriptor: | NICKEL (II) ION, YopN. Phi3T_93 | | Authors: | Zamora-Caballero, S, Marina, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.310147 Å) | | Cite: | Antagonistic interactions between phage and host factors control arbitrium lysis-lysogeny decision.
Nat Microbiol, 9, 2024
|
|
8ANV
 
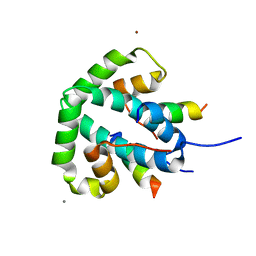 | |
4IXQ
 
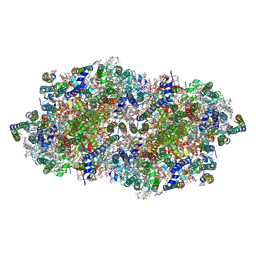 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
5GVZ
 
 | |
6CPG
 
 | | Structure of dephosphorylated Aurora A (122-403) in complex with inhibiting monobody and AT9283 in an inactive conformation | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Aurora kinase A, Monobody | | Authors: | Otten, R, Kutter, S, Zorba, A, Padua, R.A.P, Koide, A, Koide, S, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
5KAU
 
 | | The structure of SAV2435 bound to RHODAMINE 6G | | Descriptor: | GLYCEROL, RHODAMINE 6G, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
4EVU
 
 | | Crystal structure of C-terminal domain of putative periplasmic protein ydgH from S. enterica | | Descriptor: | CHLORIDE ION, Putative periplasmic protein ydgH, SULFATE ION | | Authors: | Michalska, K, Cui, H, Xu, X, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Savchenko, A, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-30 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
2HVG
 
 | | Crystal Structure of Adenylosuccinate Lyase from Plasmodium Vivax | | Descriptor: | Adenylosuccinate lyase, SULFATE ION | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Wasney, G.A, Vedadi, M, Ren, H, Alam, Z, Qiu, W, Kozieradzki, I, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
6BIO
 
 | |
5CE9
 
 | | structure of tyrosinase from walnut (Juglans regia) | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase, ... | | Authors: | Bijelic, A, Pretzler, M, Zekiri, F, Rompel, A. | | Deposit date: | 2015-07-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Plant Tyrosinase from Walnut Leaves Reveals the Importance of """"Substrate-Guiding Residues"""" for Enzymatic Specificity.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6C2R
 
 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
