8XKC
 
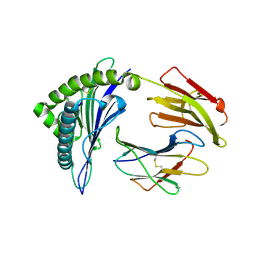 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8IK0
 
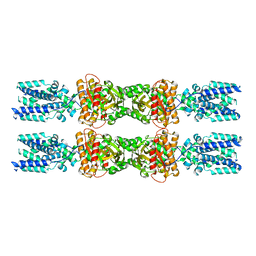 | |
8IK3
 
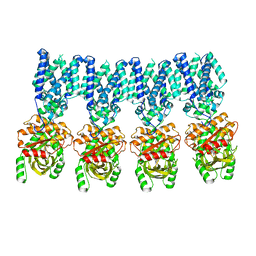 | |
7YER
 
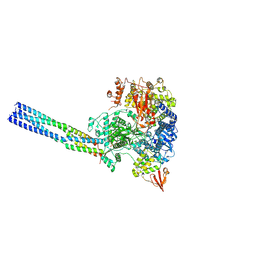 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
5ZHO
 
 | |
7YET
 
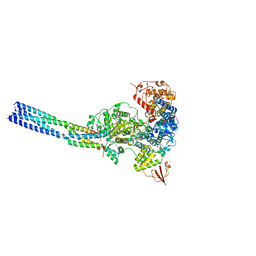 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YES
 
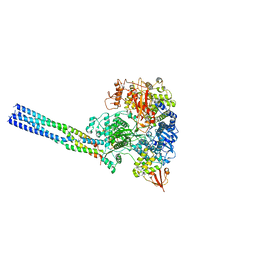 | |
8Z08
 
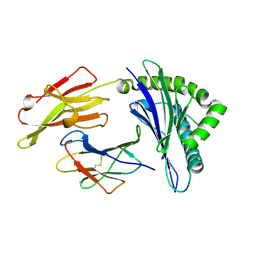 | |
8YZW
 
 | |
8XHF
 
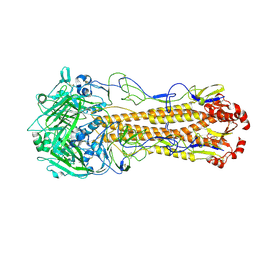 | | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 avian influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, J, Zheng, T.Y. | | Deposit date: | 2023-12-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Dual receptor-binding, infectivity, and transmissibility of an emerging H2N2 low pathogenicity avian influenza virus.
Nat Commun, 15, 2024
|
|
8ZWE
 
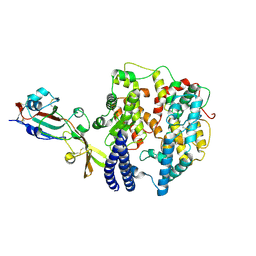 | | Cryo-EM structure of MRCoV RBD in complex with mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ji, W, Zhang, S. | | Deposit date: | 2024-06-12 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A MERS-CoV-like mink coronavirus uses ACE2 as an entry receptor.
Nature, 642, 2025
|
|
8XES
 
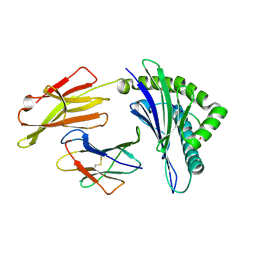 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
5ZHG
 
 | |
6J2A
 
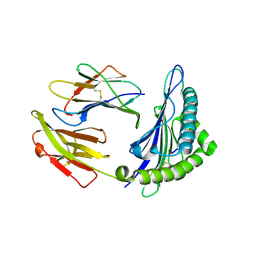 | | The structure of HLA-A*3003/NP44 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, NP44 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J9O
 
 | |
6J29
 
 | | The structure of HLA-A*3003/MTB | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, MTB | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6K7T
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1--human beta-2 microglobulin | | Descriptor: | Beta-2-microglobulin, HeV1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6K7O
 
 | | Complex structure of LILRB4 and h128-3 antibody | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, h128-3 Fab heavy chain, h128-3 Fab light chain | | Authors: | Song, H, Chai, Y, Xu, X, Gao, F.G. | | Deposit date: | 2019-06-08 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Disrupting LILRB4/APOE Interaction by an Efficacious Humanized Antibody Reverses T-cell Suppression and Blocks AML Development.
Cancer Immunol Res, 7, 2019
|
|
6K7U
 
 | | Crystal structure of beta-2 microglobulin (beta2m) of Bat (Pteropus Alecto) | | Descriptor: | Bat beta-2-microglobulin | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
8XYM
 
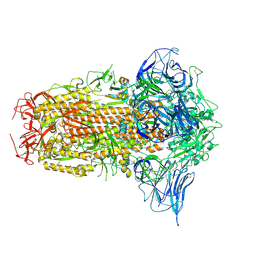 | | Cryo-EM structure of CX1 spike protein (6P) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xu, Z.P, Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-20 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural resemblances of bat-origin coronaviruses CX1 and BANAL-20-52 spikes provide further evidence on bat origination of SARS-CoV-2
To Be Published
|
|
8XYH
 
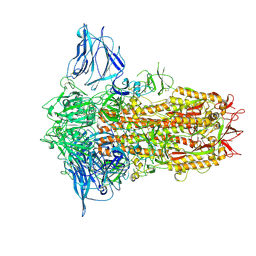 | | Cryo-EM structure of BANAL-20-52 spike protein (6P) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Xu, Z.P, Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural resemblances of bat-origin coronaviruses CX1 and BANAL-20-52 spikes provide further evidence on bat origination of SARS-CoV-2
To Be Published
|
|
8XYO
 
 | | Cryo-EM structure of CX1 receptor binding domain in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Z.P, Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-20 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural resemblances of bat-origin coronaviruses CX1 and BANAL-20-52 spikes provide further evidence on bat origination of SARS-CoV-2
To Be Published
|
|
6IW2
 
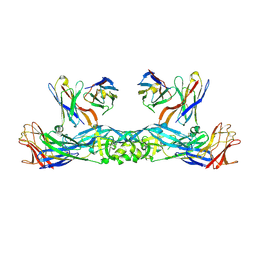 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in prefusion state | | Descriptor: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
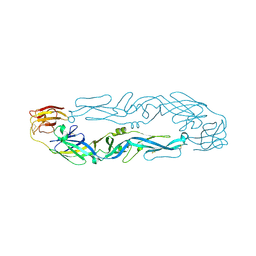
6IW5
 
 | |
6IW4
 
 | |
