7U5B
 
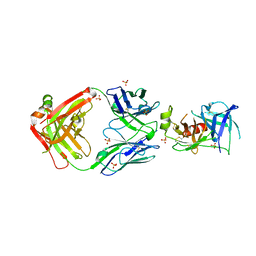 | | Structure of Human KLK5 bound to anti-KLK5 Fab | | Descriptor: | Kallikrein-5, SULFATE ION, anti-KLK5 Fab Heavy Chain, ... | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Dual antibody inhibition of KLK5 and KLK7 for Netherton syndrome and atopic dermatitis.
Sci Transl Med, 14, 2022
|
|
4ZLK
 
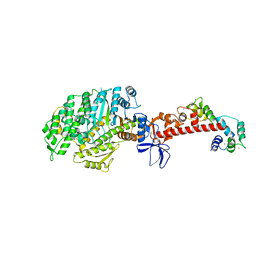 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
8X1B
 
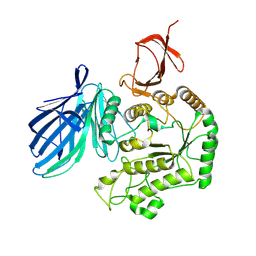 | | Cryo-EM structure of FpGalactosaminidase from Flavonifractor plautii in apo state | | Descriptor: | Alpha-galactosidase | | Authors: | Wu, G, Han, P, Su, C, Zhou, M, Luo, K. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis of FpGalNase and its combination with FpGalNAcDeAc for efficient A-to-O blood group conversion.
Exp Hematol Oncol, 14, 2025
|
|
7C9Z
 
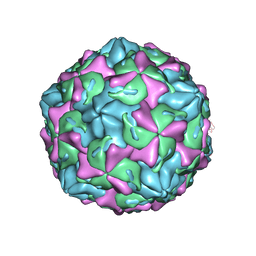 | | Coxsackievirus B1 F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Feng, R, Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CF8
 
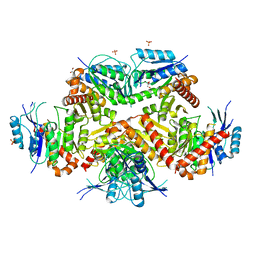 | | PfkB(Mycobacterium marinum) | | Descriptor: | Fructokinase, PfkB, GLYCEROL, ... | | Authors: | Li, J, Gao, B, Ji, R. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
8XRQ
 
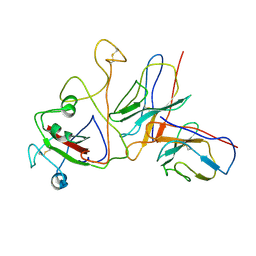 | | SARS-CoV-2 BA.1 spike RBD in complex bound with VacBB-639 | | Descriptor: | Heavy chain of VacBB 639 Fab, Light chain of VacBB 639 Fab, Spike protein S1 | | Authors: | Liu, C.C, Ju, B, Zhang, Z. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Rapid clonal expansion and somatic hypermutation contribute to the fate of SARS-CoV-2 broadly neutralizing antibodies.
J Immunol., 214, 2025
|
|
7C9X
 
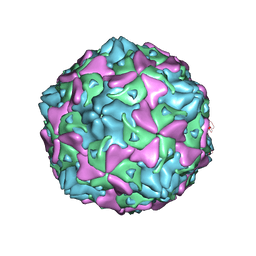 | | Echovirus 3 F-particle | | Descriptor: | MYRISTIC ACID, SPHINGOSINE, VP1, ... | | Authors: | Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9Y
 
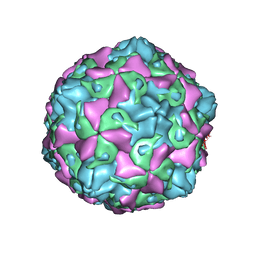 | | Coxsackievirus B5 (CVB5) F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7FCA
 
 | | PfkB(Mycobacterium marinum) | | Descriptor: | Fructokinase, PfkB, GLYCEROL, ... | | Authors: | Li, J, Gao, B, Ji, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-04 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
3TKD
 
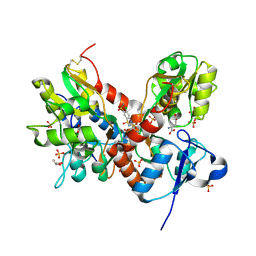 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and cyclothiazide at 1.45 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-26 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
3TDJ
 
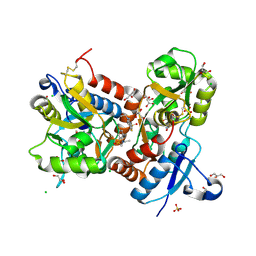 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-97 at 1.95 A resolution | | Descriptor: | 4-ethyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
2AL4
 
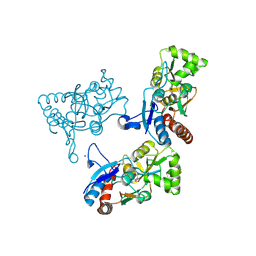 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
2AL5
 
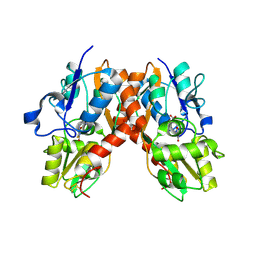 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with fluoro-willardiine and aniracetam | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
6L2H
 
 | | CGTase mutant-Y167H | | Descriptor: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | Authors: | Fan, T.W, Hou, A.Q, Chao, Y.P, Sun, Y. | | Deposit date: | 2019-10-03 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structure basis of a mutant a-CGTase tyrosine167histidine from Bacillus sp. 602-1 with enhanced a-CD production
To Be Published
|
|
2I3W
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
5CXM
 
 | | Crystal structure of the cyanobacterial plasma membrane Rieske protein PetC3 from Synechocystis PCC 6803 | | Descriptor: | Cytochrome b6/f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER, NICKEL (II) ION, ... | | Authors: | Veit, S, Takeda, K, Miki, K, Roegner, M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterisation of the cyanobacterial PetC3 Rieske protein family.
Biochim. Biophys. Acta, 1857, 2016
|
|
4YO0
 
 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 with bound PA peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antigen binding fragment, Fab, ... | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
4YNY
 
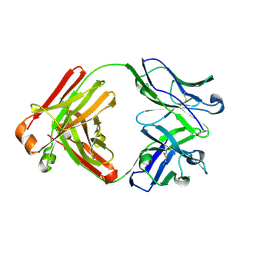 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 | | Descriptor: | Heavy chain of antigen binding fragment, Fab, Light chain of antigen binding fragment | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
3WWX
 
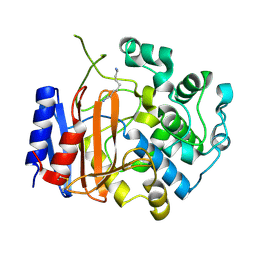 | | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 | | Descriptor: | OCTANE 1,8-DIAMINE, S12 family peptidase | | Authors: | Arima, J, Nagano, S, Hino, T, Shimone, K, Isoda, Y, Mori, N. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 - insight into the structural factors for substrate specificity.
Febs J., 283, 2016
|
|
5GUK
 
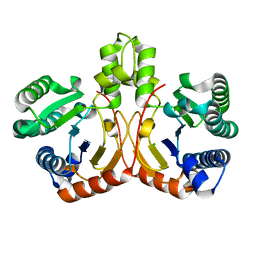 | | Crystal structure of apo form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | CHLORIDE ION, Cyclolavandulyl diphosphate synthase | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5GUL
 
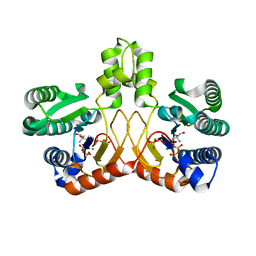 | | Crystal structure of Tris/PPix2/Mg2+ bound form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclolavandulyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1IYM
 
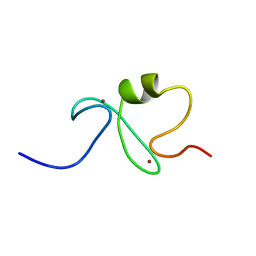 | | RING-H2 finger domain of EL5 | | Descriptor: | EL5, ZINC ION | | Authors: | Katoh, E, Katoh, S, Minami, E, Yamazaki, T. | | Deposit date: | 2002-08-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
J.Biol.Chem., 278, 2003
|
|
3AL8
 
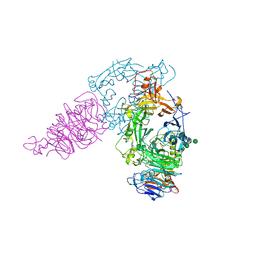 | | Plexin A2 / Semaphorin 6A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2, ... | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AL9
 
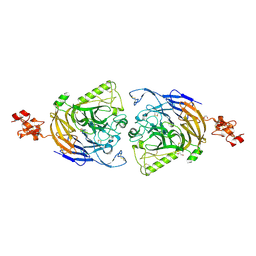 | | Mouse Plexin A2 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2 | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AFC
 
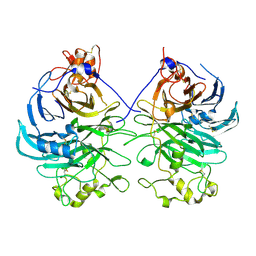 | | Mouse Semaphorin 6A extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-6A | | Authors: | Yasui, N, Nogi, T, Mihara, E, Takagi, J. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
