5GQF
 
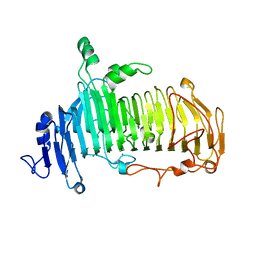 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5T8J
 
 | |
5GQC
 
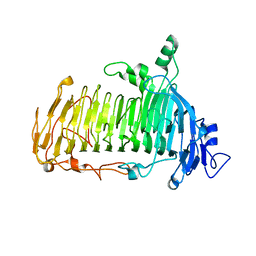 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5T8E
 
 | |
5V8Q
 
 | | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs): Part III | | Descriptor: | 4-[(2S,3S)-2-ethyl-3-hydroxy-5-oxopyrrolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, GLYCEROL | | Authors: | Wilson, K.P. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs) Part III: Discovery of 4-(5-oxopyrrolidine-1-yl)benzonitrile derivative 2f as a clinical candidate.
Bioorg. Med. Chem., 25, 2017
|
|
6MTT
 
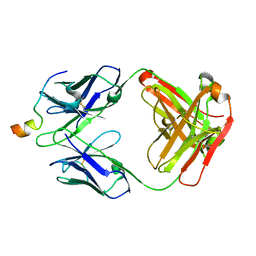 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
2KSE
 
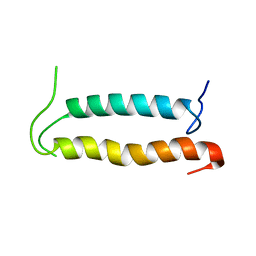 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6MTQ
 
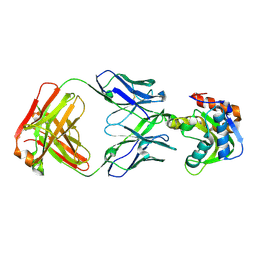 | | Crystal structure of VRC42.N1 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.N1 Fab heavy chain, Antibody VRC42.N1 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Law, W.H, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
2KSD
 
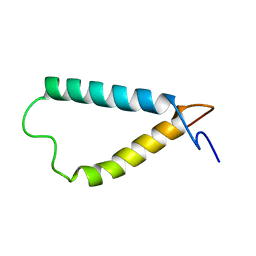 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor ArcB, Center for Structures of Membrane Proteins (CSMP) target 4310C | | Descriptor: | Aerobic respiration control sensor protein arcB | | Authors: | Maslennikov, I, Klammt, C, Hwang, E, Kefala, G, Kwiatkowski, W, Jeon, Y, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XDN
 
 | | Crystal structure of human voltage-dependent anion channel 1 (hVDAC1) in P22121 space group | | Descriptor: | DECANE, DODECANE, HEXANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structural characterization reveals novel oligomeric interactions of human voltage-dependent anion channel 1
Protein Sci., 26, 2017
|
|
7R08
 
 | | Abortive infection DNA polymerase Abi-P2 | | Descriptor: | Reverse transcriptase | | Authors: | Gapinska, M.A, Figiel, M, Czarnocki Cieciura, M, Nowotny, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R06
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Figiel, M, Nowotny, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R07
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), MAGNESIUM ION | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
8JOF
 
 | | solution-structure of Ras Binding Domain (RBD) in C-RAF | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8OVA
 
 | |
8OVE
 
 | |
7VMY
 
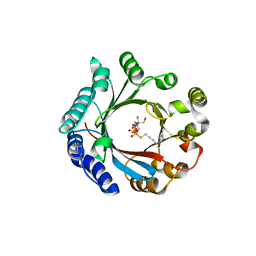 | | Crystal structure of LimF prenyltransferase bound with GSPP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
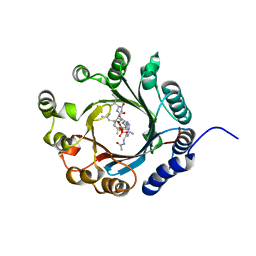 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7LVI
 
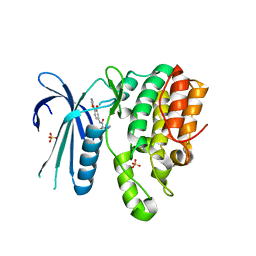 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-METHOXY-4- (1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (2R,3R)-N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-3-(propan-2-yl)piperidine-2-carboxamide, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
7LVH
 
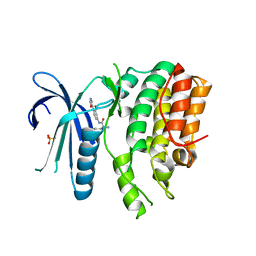 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND N-[3-METHOXY-4-(1,3-OXAZOL-5-YL)PHENYL]-3-(PROPAN-2-YL)PIPERIDINE-2-CARBOXAMIDE | | Descriptor: | 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-D-leucinamide, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
8JNA
 
 | | CRAF ras-binding domain chimera, apo form | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase, CRaf | | Authors: | Kawamura, T, Kumasaka, T. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JNB
 
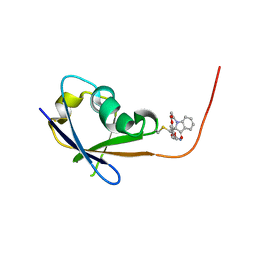 | | CRAF ras-binding domain chimera, ligand complex | | Descriptor: | 2-[4-[[(2S)-1-ethanoyl-3-oxidanylidene-2H-indol-2-yl]methyl]-2-methoxy-phenoxy]ethanamide, RAF proto-oncogene serine/threonine-protein kinase, CRaf | | Authors: | Kawamura, T, Kumasaka, T. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JOG
 
 | | solution structure of Ras Binding Domein (RBD) in C-RAF with negative allosteric modulator. | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JG5
 
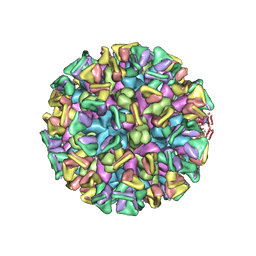 | | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | | Descriptor: | VH,SARAH, VL,SARAH, VP1 | | Authors: | Hosaka, T, Katsura, K, Kimura-Someya, T, Someya, Y, Shirouzu, M. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography revealing binding sites for human monoclonal antibodies.
J.Virol., 98, 2024
|
|
6HUS
 
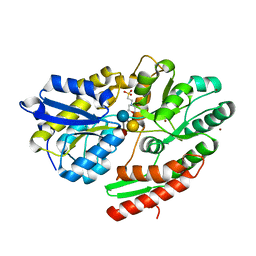 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
