7ZO7
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed cefmetazole | | Descriptor: | (2R,5R)-2-[(1S)-1-[2-(cyanomethylsulfanyl)ethanoylamino]-1-methoxy-2-oxidanyl-2-oxidanylidene-ethyl]-5-[(1-methyl-1,2,3,4-tetrazol-5-yl)sulfanylmethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
5NDE
 
 | |
5NE3
 
 | | L2 class A serine-beta-lactamase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
5MOZ
 
 | | OXA-10 Avibactam complex with bound Iodide | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, CHLORIDE ION, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
8AKI
 
 | | Acyl-enzyme complex of ampicillin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
5NE1
 
 | | L2 class A serine-beta-lactamase in complex with cyclic boronate 2 | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Beta-lactamase, TRIETHYLENE GLYCOL | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
5LRM
 
 | | Structure of di-zinc MCR-1 in P41212 space group | | Descriptor: | GLYCEROL, ZINC ION, phosphatidylethanolamine transferase Mcr-1 | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
5LRN
 
 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
6Z21
 
 | | Crystal structure of deacylation mutant KPC-2 (E166Q) | | Descriptor: | CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Natural variants modify Klebsiella pneumoniae carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance.
J.Biol.Chem., 296, 2020
|
|
6Z25
 
 | | Acylenzyme complex of ceftazidime bound to deacylation mutant KPC-4 (E166Q) | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Natural variants modify Klebsiella pneumoniae carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance.
J.Biol.Chem., 296, 2020
|
|
6Z22
 
 | |
6Z23
 
 | | Acylenzyme complex of cefotaxime bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Natural variants modify Klebsiella pneumoniae carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance.
J.Biol.Chem., 296, 2020
|
|
6Z24
 
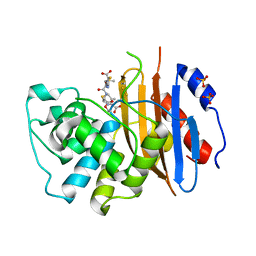 | |
7A1F
 
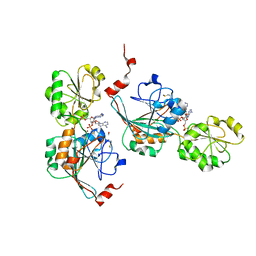 | | Crystal structure of human 5' exonuclease Appollo in complex with 5'dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5' exonuclease Apollo, FE (III) ION, ... | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
4EZH
 
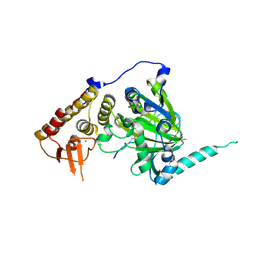 | |
4EYU
 
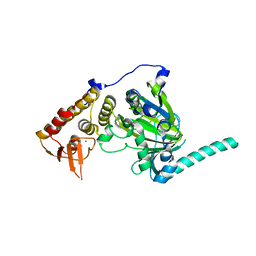 | |
4EZ4
 
 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | Descriptor: | ACETYL COENZYME *A, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2XLN
 
 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE,, SULFATE ION, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2010-07-21 | | Release date: | 2011-01-26 | | Last modified: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
2XDV
 
 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | Authors: | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-07 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2XUE
 
 | | CRYSTAL STRUCTURE OF JMJD3 | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Rowland, P, Mosley, J, Thomas, P.J. | | Deposit date: | 2010-10-19 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
2WPX
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2X71
 
 | | Structural basis for the interaction of lactivicins with serine beta- lactamases | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, BETA-LACTAMASE, ETHANOL, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2010-02-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Interaction of Lactivicins with Serine Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
2XML
 
 | | Crystal structure of human JMJD2C catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4C, ... | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Pike, A.C.W, von Delft, F, Ng, S, Carpenter, L, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
2XK1
 
 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2010-07-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
